
最新刊期
卷 26 , 期 9 , 2021
-
 摘要:Medical image registration (MIR) has aimed to implement the optimal transformation via aligning anatomical structures of a pair of medical images spatially. The crucial clinical applications like disease diagnosis, surgical guidance and radiation therapy have been envolved. Scholors have categorized MIR into inter-/intra-patient registration, uni-/multi-modal registration and rigid/non-rigid registration. Image classification has been developing deep learning-based (DL-based) MIR methods. The DL-based MIR has demonstrated substantial improvement in computational efficiency and task-specific registration accuracy over traditional iterative registration approaches. A sophisticated literature review of DL-based MIR have benefited to the disciplines. The current MIR has been analysed based on iterative optimization to one-step prediction and supervised learning to unsupervised learning. The DL-based MIR has been classified into fully supervised, dual supervised, weakly supervised and unsupervised approaches to train the DL network via the amount of supervision. Each category has been systematically reviewed. At the beginning, fully supervised methods have been reviewed in terms of the initial exploration to remove the time-consuming with low inference speed issues of deep iterative registration algorithms (deep similarity-based registration, reinforcement learning-based registration). One-step fully supervised registration has predicted the final transformation. The lack of training datasets with ground-truth transformations have barriered to train a fully supervised registration network. Most scholors have generated synthesized transformations with the following three approaches as below: 1) random augmentation-based generation; 2) traditional registration-based generation; 3) model-based generation. Next, the integration of dual-supervised and weak supervised registration have alleviated the reliance on ground truth compared with fully supervised approaches via the transition technologies between fully supervised and unsupervised methods. Dual-supervised registration frameworks have integrated image similarity metric to supervise the training. Weak supervised registration in the context of anatomical labels of interest (solid organs, vessels, ducts, structure boundaries and other subject-specific ad hoc landmarks) has replaced ground truth. The label similarity using label-driven supervised registration has facilitated the network to directly estimate the transformation for paired fixed image and moving image. The end-to-end unsupervision has been used to indicate the DL-based medical image registration evolved into the unsupervised field gradually. The unsupervision has avoided the acquisition of ground-truth transformations and segmentation labels for the supervised methods. Unsupervised registration frameworks have performed spatial data based on spatial transformer network (STN) to flat image similarity loss calculation during the training process with unknown transformations further. The latest developments and applications of DL-based unsupervised registration methods have been summarized from the aspects of loss functions and network architectures. DL-based unsupervised registration algorithms on liver CT(computed tomography) scan datasets have also been re-implemented. The demonstrated analyses have the priority to baseline model. At the end, the potentials and possibilities have been illustrated as following: 1) constructing more robust similarity metrics and more effective regularization terms to deal with multi-modality MIR; 2) quantifying registration result confidence of various DL-based models or integrating domain knowledge into current data-driven networks; 3) designing more qualified networks with fewer parameters (e.g., 3D convolution factorization, capsule network architecture).关键词:medical image registration;deep learning(DL);full supervised learning;dual supervised learning;weakly supervised learning;unsupervised learning295|430|6更新时间:2024-05-07
摘要:Medical image registration (MIR) has aimed to implement the optimal transformation via aligning anatomical structures of a pair of medical images spatially. The crucial clinical applications like disease diagnosis, surgical guidance and radiation therapy have been envolved. Scholors have categorized MIR into inter-/intra-patient registration, uni-/multi-modal registration and rigid/non-rigid registration. Image classification has been developing deep learning-based (DL-based) MIR methods. The DL-based MIR has demonstrated substantial improvement in computational efficiency and task-specific registration accuracy over traditional iterative registration approaches. A sophisticated literature review of DL-based MIR have benefited to the disciplines. The current MIR has been analysed based on iterative optimization to one-step prediction and supervised learning to unsupervised learning. The DL-based MIR has been classified into fully supervised, dual supervised, weakly supervised and unsupervised approaches to train the DL network via the amount of supervision. Each category has been systematically reviewed. At the beginning, fully supervised methods have been reviewed in terms of the initial exploration to remove the time-consuming with low inference speed issues of deep iterative registration algorithms (deep similarity-based registration, reinforcement learning-based registration). One-step fully supervised registration has predicted the final transformation. The lack of training datasets with ground-truth transformations have barriered to train a fully supervised registration network. Most scholors have generated synthesized transformations with the following three approaches as below: 1) random augmentation-based generation; 2) traditional registration-based generation; 3) model-based generation. Next, the integration of dual-supervised and weak supervised registration have alleviated the reliance on ground truth compared with fully supervised approaches via the transition technologies between fully supervised and unsupervised methods. Dual-supervised registration frameworks have integrated image similarity metric to supervise the training. Weak supervised registration in the context of anatomical labels of interest (solid organs, vessels, ducts, structure boundaries and other subject-specific ad hoc landmarks) has replaced ground truth. The label similarity using label-driven supervised registration has facilitated the network to directly estimate the transformation for paired fixed image and moving image. The end-to-end unsupervision has been used to indicate the DL-based medical image registration evolved into the unsupervised field gradually. The unsupervision has avoided the acquisition of ground-truth transformations and segmentation labels for the supervised methods. Unsupervised registration frameworks have performed spatial data based on spatial transformer network (STN) to flat image similarity loss calculation during the training process with unknown transformations further. The latest developments and applications of DL-based unsupervised registration methods have been summarized from the aspects of loss functions and network architectures. DL-based unsupervised registration algorithms on liver CT(computed tomography) scan datasets have also been re-implemented. The demonstrated analyses have the priority to baseline model. At the end, the potentials and possibilities have been illustrated as following: 1) constructing more robust similarity metrics and more effective regularization terms to deal with multi-modality MIR; 2) quantifying registration result confidence of various DL-based models or integrating domain knowledge into current data-driven networks; 3) designing more qualified networks with fewer parameters (e.g., 3D convolution factorization, capsule network architecture).关键词:medical image registration;deep learning(DL);full supervised learning;dual supervised learning;weakly supervised learning;unsupervised learning295|430|6更新时间:2024-05-07 -
 摘要:Medical imaging has been a proactive tool for doctors to diagnose and treat diseases via the qualitative and quantitative analyses based on non-invasive lesions. Medical images have been interpreted via computer tomography (CT), X-ray, magnetic resonance imaging (MRI) and positron emission tomography (PET). The barriers of medical image segmentation need to be resolved due to low contrast amongst the lesion, the surrounding tissue and blurred edges of the lesion. Labeling manually for hundreds of slices of organs or lesions has been quite time-consuming due to anatomy of the human body and shape of lesions. Manual labeling has intended to high subjective and low reproducibility. Doctors have been beneficial from a automatically locating, segmenting and quantifying lesions. Deep learning has been used widely in medical image processing. Deep learning-based U-Net has played a key role in the lesions segmentation. The encoding and decoding ways has made U-Net structures simply and symmetrically. Features extraction of medical images has been realized via convolution and down-sampling operations. The image segmentation mask via the transposed convolution and concatenation has been interpreted. A small-sized dataset has achieved qualified medical image segmentation. U-Net has been summarized and analyzed on the four aspects: the definition of U-Net, the upgrading of U-Net model, the setup of U-Net structure and the mechanism of U-Net. Four research areas have been proposed as below: 1) the basic structure and working principle of U-Net via convolution operation, down sampling, up sampling and concatenation. 2) U-Net network model have been demonstrated in three aspects in the context of the number of encoders, multiple U-Net cascades and other models combined with U-Net. U-Net based network have been divided into two, three and four encoders further in terms of the amount of encoders: Y-Net, Ψ-Net and multi-path dense U-Net. Multiple U-Nets cascade has been categorized into multiple U-Nets in series and multiple U-Nets in parallel based on the cascades mode of multiple U-Nets. In addition U-Net has improved the segmentation performance on the aspects of dual tree complex wavelet transform, local difference method, level set, random walk, graph cutting, CNNs(convolutional neural networks) and deep reinforcement learning. The upgrading of U-Net network structure have been divided into six subcategories including image augmentation, convolution operation, down-sampling operation, up-sampling operation, model optimization strategies and concatenation. Image enhancement has be divided into elastic deformation, geometric transformation, generative adversarial networks (GAN), Wasserstein generative adversarial networks (WGAN) and real-time image enhancement further. The convolution operation has been improved via padding mode and convolution redesign. The padding mode mentioned has adapted constant padding, zero padding, replication padding and reflection padding and improvements to dilated convolution, inception module and asymmetric convolution. The down-sampling has been improved via max-pooling, average-pooling, stride convolution, dilated convolution, inception module and spatial pyramid pooling. Several up-sampling improvements have illustrated simultaneously via sub-pixel convolution, transposed convolution, nearest neighbor interpolation, bilinear interpolation and trilinear interpolation. Model optimization strategies have been divided into two aspects in detail of activation function and normalization, the improvements of activation function includes rectified linear unit(ReLU), parametric ReLU(PReLU), random ReLU(RReLU), leaky ReLU(LReLU), hard exponential linear sigmoid squahing(HardELiSH) and exponential linear sigmoid squashing(ELiSH), and normalization method. The improvements have been to shown based on batch normalization, group normalization, instance normalization and layer normalization. The concatenation based improvement has been one of the future research area. The current concatenation improvements have been mainly realized via attention mechanism, new concatenation, feature reuse and de-convolution with activation function, annotation information fusion from Siamese network. The improved mechanisms in the U-Net network have been emphasized based on residual mechanism, dense mechanism, attention mechanism and the multi-mechanisms integration. The segmentation performance of the network can be enhanced. The further four research areas in U-Net have been illustrated as below: 1) the generalization of deep learning methods cannot be customized to fit the segmentation network for specific scenarios in the future. 2) Supervised deep learning models have required a lot of annotated images labeled for treatment. Unsupervised and semi-supervised deep learning models have been a vital research work further. 3) The low interpretability of U-Net network has lead the low acceptance in the mechanism of its operation.4) More accurate segmentation mask with fewer parameters has been obtained via good quality network structure. The precise manual segmentation has been so time-consuming and labor intensive. The simplified and quick semi-automatic segmentation has relied on the parameters and user-specified image preprocessing. The deep learning-based U-Net network has been segmented the lesions quickly, accurately and consistently. The structure, improvements and further research areas of U-Net network have been analyzed to the development of U-Net network.关键词:U-Net;medical image;semantic segmentation;network structure;network model209|158|27更新时间:2024-05-07
摘要:Medical imaging has been a proactive tool for doctors to diagnose and treat diseases via the qualitative and quantitative analyses based on non-invasive lesions. Medical images have been interpreted via computer tomography (CT), X-ray, magnetic resonance imaging (MRI) and positron emission tomography (PET). The barriers of medical image segmentation need to be resolved due to low contrast amongst the lesion, the surrounding tissue and blurred edges of the lesion. Labeling manually for hundreds of slices of organs or lesions has been quite time-consuming due to anatomy of the human body and shape of lesions. Manual labeling has intended to high subjective and low reproducibility. Doctors have been beneficial from a automatically locating, segmenting and quantifying lesions. Deep learning has been used widely in medical image processing. Deep learning-based U-Net has played a key role in the lesions segmentation. The encoding and decoding ways has made U-Net structures simply and symmetrically. Features extraction of medical images has been realized via convolution and down-sampling operations. The image segmentation mask via the transposed convolution and concatenation has been interpreted. A small-sized dataset has achieved qualified medical image segmentation. U-Net has been summarized and analyzed on the four aspects: the definition of U-Net, the upgrading of U-Net model, the setup of U-Net structure and the mechanism of U-Net. Four research areas have been proposed as below: 1) the basic structure and working principle of U-Net via convolution operation, down sampling, up sampling and concatenation. 2) U-Net network model have been demonstrated in three aspects in the context of the number of encoders, multiple U-Net cascades and other models combined with U-Net. U-Net based network have been divided into two, three and four encoders further in terms of the amount of encoders: Y-Net, Ψ-Net and multi-path dense U-Net. Multiple U-Nets cascade has been categorized into multiple U-Nets in series and multiple U-Nets in parallel based on the cascades mode of multiple U-Nets. In addition U-Net has improved the segmentation performance on the aspects of dual tree complex wavelet transform, local difference method, level set, random walk, graph cutting, CNNs(convolutional neural networks) and deep reinforcement learning. The upgrading of U-Net network structure have been divided into six subcategories including image augmentation, convolution operation, down-sampling operation, up-sampling operation, model optimization strategies and concatenation. Image enhancement has be divided into elastic deformation, geometric transformation, generative adversarial networks (GAN), Wasserstein generative adversarial networks (WGAN) and real-time image enhancement further. The convolution operation has been improved via padding mode and convolution redesign. The padding mode mentioned has adapted constant padding, zero padding, replication padding and reflection padding and improvements to dilated convolution, inception module and asymmetric convolution. The down-sampling has been improved via max-pooling, average-pooling, stride convolution, dilated convolution, inception module and spatial pyramid pooling. Several up-sampling improvements have illustrated simultaneously via sub-pixel convolution, transposed convolution, nearest neighbor interpolation, bilinear interpolation and trilinear interpolation. Model optimization strategies have been divided into two aspects in detail of activation function and normalization, the improvements of activation function includes rectified linear unit(ReLU), parametric ReLU(PReLU), random ReLU(RReLU), leaky ReLU(LReLU), hard exponential linear sigmoid squahing(HardELiSH) and exponential linear sigmoid squashing(ELiSH), and normalization method. The improvements have been to shown based on batch normalization, group normalization, instance normalization and layer normalization. The concatenation based improvement has been one of the future research area. The current concatenation improvements have been mainly realized via attention mechanism, new concatenation, feature reuse and de-convolution with activation function, annotation information fusion from Siamese network. The improved mechanisms in the U-Net network have been emphasized based on residual mechanism, dense mechanism, attention mechanism and the multi-mechanisms integration. The segmentation performance of the network can be enhanced. The further four research areas in U-Net have been illustrated as below: 1) the generalization of deep learning methods cannot be customized to fit the segmentation network for specific scenarios in the future. 2) Supervised deep learning models have required a lot of annotated images labeled for treatment. Unsupervised and semi-supervised deep learning models have been a vital research work further. 3) The low interpretability of U-Net network has lead the low acceptance in the mechanism of its operation.4) More accurate segmentation mask with fewer parameters has been obtained via good quality network structure. The precise manual segmentation has been so time-consuming and labor intensive. The simplified and quick semi-automatic segmentation has relied on the parameters and user-specified image preprocessing. The deep learning-based U-Net network has been segmented the lesions quickly, accurately and consistently. The structure, improvements and further research areas of U-Net network have been analyzed to the development of U-Net network.关键词:U-Net;medical image;semantic segmentation;network structure;network model209|158|27更新时间:2024-05-07 -
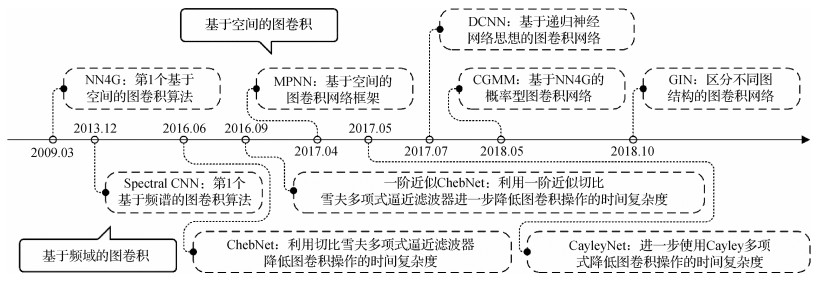 摘要:The convolutional neural networks (CNN) have been facilitated to develop deep learning-based medical image sustainable research. The translation invariance capability has constrained the expression of CNN in the context of non-Euclidean spatial data. In order to realize deep learning-based spatial feature extraction, graph convolution has resolved the topology modeling issue based on non-Euclidean spatial data. The latest theories and applications of graph convolutional networks (GCN) for medical image analysis have been reviewed. This research has been divided into four aspects as follows: 1) Data structure transformation of medical images based on graph-structure; 2) Theoretical development and network architecture of GCN; 3) The optimized and derivative of graph convolution mechanism; 4) GCN implementation in medical image segmentation, disease detection, and image reconstruction. First, graph-structure-based medical images transformation has been reviewed in the context of graph data acquisition, transformation, and reconstruction. The graph-structure-based medical data have been acquired via the professional medical equipment, the sparse pruning algorithm, or the rebuilt graph-structure using the K-nearest neighbor (KNN) algorithm. The graph-structure reconstruction algorithm based on the medical image features has performed better than the graph-structure conversion algorithm based on the medical image data. Next, the critical architecture of the GCN, including the graph convolutional layer, the graph regularization layer, the graph pooling layer, and the graph readout layer, has been summarized. The graph-structural nodes or edges have been updated via the graph convolution layer. The generalization of GCN has been upgraded via the graph regularization layer. The number of calculation parameters has been reduced via the graph pooling layer. The representation of the graph has been generated via the graph readout layer. Graph convolution has been categorized into two methods as mentioned below: a) The spectrum-based graph convolution operation has been implemented via the theory of graph spectrum; b) The spatial domain-based graph convolution operation has been defined via the connectivity of each node. The spectrum-based graph convolution has relied on the eigen-decomposition of the Laplace matrix with the defects of high time complexity, poor portability, and narrow application. The convolution can be optimized by Chebyshev Inequality analysis. The graph pooling layer has effectively reduced parameter size. The graph regularization layer can facilitate the generalization of the model and alleviate the over-fitting and over-smoothing issues. The different structural features, node features, and edge features have been extracted based on the graph convolutional layer. All features need to be aggregated to complete the classification (note: this operation is called the readout operation, and its function is similar to the fully connected layer of CNN). Third, the development and derivation mechanism of GCN optimizations have been summarized. For instance, the jump connection mechanism of deepGCN has alleviated the over-smooth issue. The outputs of multiple GCN based on inception architecture can be integrated to improve the representation ability of the model. The graph attention mechanism has aggregated the differentiated information of the GCN nodes. The adjacency matrix reconstruction has been critically optimized to achieve qualified GCN model performance via learning the hidden structure of the unidentified graph adjacency matrix. Fourth, the main application of GCN for medical image analysis has been interpreted. The general graph-structure construct algorithm for GCN application to medical image segmentation has taken the region of interest (ROI) as the node and the existence of connection in the ROI as the edge. For some unique imaging data (such as brain voxel data and cardiac coronary artery surface grid data), the KNN algorithm has been used to convert them into a graph-structure. The improvement of model architecture has changed from the simple stack of CNN and GCN to the complex combination of various models. The previous medical images application of GCN in disease detection has mainly focused on brain images. Disease detection has been accomplished by using GCN based on the various relationships between objects. Current research on disease detection has mainly divided into three aspects: 1) various CNN models have been used to extract the features based on the original medical images; 2) the KNN algorithm or graph attention algorithm has been used for feature reconstruction; 3) the potential relationship between features is mined by graph convolution for feature classification. In addition, GCN have been used for brain magnetic resonance imaging (MRI) reconstruction, liver image reconstruction, heart image reconstruction, and other diagnoses. In a word, GCN have effectively mined the generalized topological structure in image data on the aspects of medical image segmentation, disease detection, and image reconstruction. The integrated deep learning architecture, which uses pre-trained CNN as feature extractor and GCN as the classifier, has solved the missing issues of medical training samples in a graph structure and significantly improved the performance of deep learning technology in medical image analysis.关键词:medical image;deep learning;graph representation learning;graph neural network(GNN);graph convolution network(GCN)230|150|5更新时间:2024-05-07
摘要:The convolutional neural networks (CNN) have been facilitated to develop deep learning-based medical image sustainable research. The translation invariance capability has constrained the expression of CNN in the context of non-Euclidean spatial data. In order to realize deep learning-based spatial feature extraction, graph convolution has resolved the topology modeling issue based on non-Euclidean spatial data. The latest theories and applications of graph convolutional networks (GCN) for medical image analysis have been reviewed. This research has been divided into four aspects as follows: 1) Data structure transformation of medical images based on graph-structure; 2) Theoretical development and network architecture of GCN; 3) The optimized and derivative of graph convolution mechanism; 4) GCN implementation in medical image segmentation, disease detection, and image reconstruction. First, graph-structure-based medical images transformation has been reviewed in the context of graph data acquisition, transformation, and reconstruction. The graph-structure-based medical data have been acquired via the professional medical equipment, the sparse pruning algorithm, or the rebuilt graph-structure using the K-nearest neighbor (KNN) algorithm. The graph-structure reconstruction algorithm based on the medical image features has performed better than the graph-structure conversion algorithm based on the medical image data. Next, the critical architecture of the GCN, including the graph convolutional layer, the graph regularization layer, the graph pooling layer, and the graph readout layer, has been summarized. The graph-structural nodes or edges have been updated via the graph convolution layer. The generalization of GCN has been upgraded via the graph regularization layer. The number of calculation parameters has been reduced via the graph pooling layer. The representation of the graph has been generated via the graph readout layer. Graph convolution has been categorized into two methods as mentioned below: a) The spectrum-based graph convolution operation has been implemented via the theory of graph spectrum; b) The spatial domain-based graph convolution operation has been defined via the connectivity of each node. The spectrum-based graph convolution has relied on the eigen-decomposition of the Laplace matrix with the defects of high time complexity, poor portability, and narrow application. The convolution can be optimized by Chebyshev Inequality analysis. The graph pooling layer has effectively reduced parameter size. The graph regularization layer can facilitate the generalization of the model and alleviate the over-fitting and over-smoothing issues. The different structural features, node features, and edge features have been extracted based on the graph convolutional layer. All features need to be aggregated to complete the classification (note: this operation is called the readout operation, and its function is similar to the fully connected layer of CNN). Third, the development and derivation mechanism of GCN optimizations have been summarized. For instance, the jump connection mechanism of deepGCN has alleviated the over-smooth issue. The outputs of multiple GCN based on inception architecture can be integrated to improve the representation ability of the model. The graph attention mechanism has aggregated the differentiated information of the GCN nodes. The adjacency matrix reconstruction has been critically optimized to achieve qualified GCN model performance via learning the hidden structure of the unidentified graph adjacency matrix. Fourth, the main application of GCN for medical image analysis has been interpreted. The general graph-structure construct algorithm for GCN application to medical image segmentation has taken the region of interest (ROI) as the node and the existence of connection in the ROI as the edge. For some unique imaging data (such as brain voxel data and cardiac coronary artery surface grid data), the KNN algorithm has been used to convert them into a graph-structure. The improvement of model architecture has changed from the simple stack of CNN and GCN to the complex combination of various models. The previous medical images application of GCN in disease detection has mainly focused on brain images. Disease detection has been accomplished by using GCN based on the various relationships between objects. Current research on disease detection has mainly divided into three aspects: 1) various CNN models have been used to extract the features based on the original medical images; 2) the KNN algorithm or graph attention algorithm has been used for feature reconstruction; 3) the potential relationship between features is mined by graph convolution for feature classification. In addition, GCN have been used for brain magnetic resonance imaging (MRI) reconstruction, liver image reconstruction, heart image reconstruction, and other diagnoses. In a word, GCN have effectively mined the generalized topological structure in image data on the aspects of medical image segmentation, disease detection, and image reconstruction. The integrated deep learning architecture, which uses pre-trained CNN as feature extractor and GCN as the classifier, has solved the missing issues of medical training samples in a graph structure and significantly improved the performance of deep learning technology in medical image analysis.关键词:medical image;deep learning;graph representation learning;graph neural network(GNN);graph convolution network(GCN)230|150|5更新时间:2024-05-07 -
 摘要:Medical images can be classified as anatomical images and functional images. The high resolution in anatomical images can provide information about the anatomical structure and morphology of human body organs but cannot reflect the functional information of organs. There is some information about the metabolic functions of organs in functional images; however, it has low resolution and cannot display the anatomical details of organs or lesions. With the continuous development of medical imaging technology, how to use medical images comprehensively to avoid the shortcomings of different medical imaging modalities has become a complex problem. Image fusion has aimed to fuse value-added information from two images to one image in terms of the constraints of single sensor. Effective diagnose and treat diseases have been beneficial to acquire qualified fusion image of organs and tissues and lesions simultaneously via fusing positron emission tomography(PET) images and computed tomography images(CT). Research on pixel-level image fusion based on multi-scale transformation has been focused nowadays. A fusion image with rich edge information has been easy use under the circumstances of image registration. The detailed information for image fusion has been obtained via moderate image decomposition option and fusion strategical implementation. Pixel-level image fusion based on multi-scale transformation has been prior to medical image processing, video surveillance image processing, remote sensing image processing and other tasks. Pixel-level image fusion based on multi-scale transformation has been analyzed and summarized on the following five aspects: 1) mechanism of multi-scale image fusion transformation; 2) multiscale decomposition analysis; 3) a consensus pixel-level fusion framework; 4) subband fusion rules under high and low frequency; and 5) pixel-level image fusion based on multi-scale transformation application in medical image processing. First, the mechanism and framework of image fusion based on multi-scale transformation have been proposed. Next, the multi-temporal decomposition have been summarized via the pyramid decomposition evolvement, wavelet transform and multi-scale geometric analysis. Such as, the advantages of pyramid decomposition are simple implementation and fast calculation speed. The disadvantage of pyramid decomposition is that it has no directionality, sensitive to noise, has no stability during reconstruction, and there is redundancy between the layers of the pyramid. Wavelet transform does not have direction selectivity and shift invariance. The advantage of multi-scale geometric analysis is that it has multiple directions. The original image is decomposed into high-frequency subbands in different directions by nonsubsampled contourlet transform(NSCT), which can enhance the edge details of the image from many directions. Then, two consensus pixel-level fusion frameworks has been illustrated. The Zhang framework has demonstrated four analyses including activity level measurement, coefficient grouping method, coefficient combination method and consistency verification. The Piella framework has consisted of four ways on the aspects of matching measurement, activity measurement, decision module and synthesis module. Low frequency fused sub bands and high frequency fused sub-bands have been categorized. The low frequency fused sub band has been subdivided into five categories: pixel based, region based, fuzzy theory based, sparse representation based and focus metric based. Pixel-based fusion rules include average fusion rules, coefficient maximum fusion rules, etc., region-based fusion rules include regional variance maximum fusion rules, and local energy maximum fusion rules, etc., fuzzy theory fusion rules include fuzzy inference system fusion rules, etc. Fusion rules based on sparse representation include new sum of modified Laplacian, and extended sum of modified Laplacian and other fusion rules. Fusion rules based on focal measures include spatial frequency fusion rules, and local spatial frequencies fusion rules, etc. The high frequency fused sub band rules have been subdivided into five categories: pixel based, edge based, region based, sparse representation based and neural network based. Edge-based fusion rules include edge strength maximum fusion rules and guided filter-based fusion rules, etc., region-based fusion rules include average gradient fusion rules and improved average gradient fusion rules, etc., fusion rules based on sparse representation include multi-scale convolution sparse representation fusion rules and separable dictionary learning method fusion rules, etc. Fusion rules based on neural networks include parameter adaptive dual-channel pulse-coupled neural network, and adaptive dual-channel pulse-coupled neural network fusion rules, simplified pulse-coupled neural network and other fusion rules. At last, 12 pixel-level image fusion based on multi-scale transformation have been summarized for multi-modal medical image fusions. Two-mode medical image fusion and three-mode medical image fusion have been identified. The two-mode medical image fusion has been subdivided into 11 categories. The CT/magnetic resonance image(MRI)/PET has been analyzed in three-mode medical image fusion. At last, the further 4 research areas of pixel-level image fusion based on multi-scale transformation have been summarized: 1) accurate registration of image preprocessing issue; 2) fusion of ultrasonic images with other multimodal medical images issue; 3) integration of multi-algorithms for cascading decomposition; 4) multi-scale decomposition methods and fusion rules for pixel-level image fusion and the application of multi-scale transformation in medical image fusion have been summarized. This article systematically summarizes the multi-scale decomposition methods and fusion rules in the pixel-level image fusion process based on multi-scale transformation, as well as the application of multi-scale transformation in medical image fusion, and the research on the pixel-level medical image fusion method based on multi-scale transformation has a positive guiding significance.关键词:multi-scale transformation;pixel-level fusion;medical images;Zhang framework;Piella framework217|94|6更新时间:2024-05-07
摘要:Medical images can be classified as anatomical images and functional images. The high resolution in anatomical images can provide information about the anatomical structure and morphology of human body organs but cannot reflect the functional information of organs. There is some information about the metabolic functions of organs in functional images; however, it has low resolution and cannot display the anatomical details of organs or lesions. With the continuous development of medical imaging technology, how to use medical images comprehensively to avoid the shortcomings of different medical imaging modalities has become a complex problem. Image fusion has aimed to fuse value-added information from two images to one image in terms of the constraints of single sensor. Effective diagnose and treat diseases have been beneficial to acquire qualified fusion image of organs and tissues and lesions simultaneously via fusing positron emission tomography(PET) images and computed tomography images(CT). Research on pixel-level image fusion based on multi-scale transformation has been focused nowadays. A fusion image with rich edge information has been easy use under the circumstances of image registration. The detailed information for image fusion has been obtained via moderate image decomposition option and fusion strategical implementation. Pixel-level image fusion based on multi-scale transformation has been prior to medical image processing, video surveillance image processing, remote sensing image processing and other tasks. Pixel-level image fusion based on multi-scale transformation has been analyzed and summarized on the following five aspects: 1) mechanism of multi-scale image fusion transformation; 2) multiscale decomposition analysis; 3) a consensus pixel-level fusion framework; 4) subband fusion rules under high and low frequency; and 5) pixel-level image fusion based on multi-scale transformation application in medical image processing. First, the mechanism and framework of image fusion based on multi-scale transformation have been proposed. Next, the multi-temporal decomposition have been summarized via the pyramid decomposition evolvement, wavelet transform and multi-scale geometric analysis. Such as, the advantages of pyramid decomposition are simple implementation and fast calculation speed. The disadvantage of pyramid decomposition is that it has no directionality, sensitive to noise, has no stability during reconstruction, and there is redundancy between the layers of the pyramid. Wavelet transform does not have direction selectivity and shift invariance. The advantage of multi-scale geometric analysis is that it has multiple directions. The original image is decomposed into high-frequency subbands in different directions by nonsubsampled contourlet transform(NSCT), which can enhance the edge details of the image from many directions. Then, two consensus pixel-level fusion frameworks has been illustrated. The Zhang framework has demonstrated four analyses including activity level measurement, coefficient grouping method, coefficient combination method and consistency verification. The Piella framework has consisted of four ways on the aspects of matching measurement, activity measurement, decision module and synthesis module. Low frequency fused sub bands and high frequency fused sub-bands have been categorized. The low frequency fused sub band has been subdivided into five categories: pixel based, region based, fuzzy theory based, sparse representation based and focus metric based. Pixel-based fusion rules include average fusion rules, coefficient maximum fusion rules, etc., region-based fusion rules include regional variance maximum fusion rules, and local energy maximum fusion rules, etc., fuzzy theory fusion rules include fuzzy inference system fusion rules, etc. Fusion rules based on sparse representation include new sum of modified Laplacian, and extended sum of modified Laplacian and other fusion rules. Fusion rules based on focal measures include spatial frequency fusion rules, and local spatial frequencies fusion rules, etc. The high frequency fused sub band rules have been subdivided into five categories: pixel based, edge based, region based, sparse representation based and neural network based. Edge-based fusion rules include edge strength maximum fusion rules and guided filter-based fusion rules, etc., region-based fusion rules include average gradient fusion rules and improved average gradient fusion rules, etc., fusion rules based on sparse representation include multi-scale convolution sparse representation fusion rules and separable dictionary learning method fusion rules, etc. Fusion rules based on neural networks include parameter adaptive dual-channel pulse-coupled neural network, and adaptive dual-channel pulse-coupled neural network fusion rules, simplified pulse-coupled neural network and other fusion rules. At last, 12 pixel-level image fusion based on multi-scale transformation have been summarized for multi-modal medical image fusions. Two-mode medical image fusion and three-mode medical image fusion have been identified. The two-mode medical image fusion has been subdivided into 11 categories. The CT/magnetic resonance image(MRI)/PET has been analyzed in three-mode medical image fusion. At last, the further 4 research areas of pixel-level image fusion based on multi-scale transformation have been summarized: 1) accurate registration of image preprocessing issue; 2) fusion of ultrasonic images with other multimodal medical images issue; 3) integration of multi-algorithms for cascading decomposition; 4) multi-scale decomposition methods and fusion rules for pixel-level image fusion and the application of multi-scale transformation in medical image fusion have been summarized. This article systematically summarizes the multi-scale decomposition methods and fusion rules in the pixel-level image fusion process based on multi-scale transformation, as well as the application of multi-scale transformation in medical image fusion, and the research on the pixel-level medical image fusion method based on multi-scale transformation has a positive guiding significance.关键词:multi-scale transformation;pixel-level fusion;medical images;Zhang framework;Piella framework217|94|6更新时间:2024-05-07
Review
-
 摘要:ObjectiveImages-based segmentation of pulmonary anatomy has been set up the anatomical structures to formulate rapid and targeted diagnostic information. The purpose of pulmonary anatomy segmentation has been associated to a pixel in an image with an anatomical structure without the need for manual initialization. A lots of supervised deep learning image segmentation have been illustrated for segmenting regions of interest in pulmonary CT(computer tomography) images. The medical image segmentation has greatly relied on high-quality labeled medical image data, CT images-based lung anatomy labeled data has been insufficient adopted due to the lack of expert annotation of regions of interest and the lack of infrastructure and standards for sharing labeled data. Most of pulmonary CT annotation datasets have focused on thoracic cancer, pulmonary nodules, tuberculosis, pneumonia and lung segmentation. A dataset of pulmonary CT/CTA(computer tomography/computer tomography angiography) scan images with labels has facilitated the evolvement of pulmonary anatomical structure segmentation algorithms. The dataset has been evaluated the performance of state-of-the-art pulmonary anatomy structure segmentation methods for chest CT scans. It has been difficult to compare various algorithms for pulmonary anatomy structure segmentation. Different methods have been evaluated on different datasets using different evaluation measures in common. The related dataset has implemented a dataset of chest CT scans to identify varying abnormalities based on the reference standards in the context of airway, lung parenchyma, lobe and pulmonary artery. The vein segmentations have been established. The dataset has a unique calculation to compare pulmonary anatomy structure algorithms via the comparison all methods against the reference standard baseline.MethodA sum of 67 sets of CT/CTA images of the pulmonary have labeled in this dataset including 24 sets of CT images and 43 sets of CTA images via a total of 26 157 slices images. Each set of CT/CTA images have labeled for airway, lung parenchyma, lobe, pulmonary artery and vein. Multi-channel images have represented a variety of regular-based clinical scanners based on a reconstructed mediastinal window algorithm. The medical image-based dataset has been annotated and verified via. Manual corrections have annotated using internal software funded by the Key Laboratory of Medical Imaging Intelligent Computing, Ministry of Education.ResultPart of dataset representative segmentation tasks have been used via pulmonary CT anatomical structure segmentation (conference details: the medical image challenge competition held during the 4th International Symposium on Image Computing and Digital Medicine (ISICDM) in Shenyang, China). The representative dataset has included 10 groups of CT and CTA in the training dataset and 5 groups of test dataset. The challenge competition has also offered a platform for evaluating model performance of pulmonary blood vessels, airways and lung parenchyma. The result of segmentation and the effect of 3D reconstruction have evaluated by Dice coefficients, over-segmentation rate, under-segmentation rate and medical and algorithmic industry experts.ConclusionFour parts of labeled image datasets have been used as a pulmonary CT dataset. This dataset has labeled using different colored pixels and saved respectively for different pulmonary anatomies structure. The annotated data have been re-formatted to ensure easy access. The location of the markers in color pixels has been displayed via 25 000 labeled sliced images dataset using the image format of the raw data. All annotated images from the digital imaging and communications in medicine (DICOM) format to portable network graphics (PNG) images has been converted based on standard DICOM data. The chest CT image dataset has provided valid annotated data via DICOM-based sensitive information re-movement. First each set of CT/CTA has labeled with 4 different target region categories in the context of airway, lung parenchyma, lobe and pulmonary artery and vein to complement the anatomical structure of CT/CTA image dataset of the pulmonary. Next, a partially representative dataset has been and the have verified by the challenge competition. Lastly, clear and intuitive 3-dimensional visualized structural images have been reconstructed for the acquisition of each anatomical structure of the pulmonary segmented via CT/CTA images to assist in the diagnosis of pulmonary diseases. First, this dataset has not annotated lung segments. It has been difficult to obtain that the invisibility of lung segment boundaries based on targeted and accurate reference segmentation criteria. Second, the annotation data have been basically carried out on healthy images and rarely on lesion images. The most important feature of medical datasets has upgraded the diversity of data. The robustness of image segmentation have been implementing further. Last, manual annotation of medical anatomical structure images have inevitably resulted some errors. Supplementing lung segments markers and improving the diversity of data based on pulmonary CT anatomical structure segmentation algorithms have been implementing further via labeling more lesion images.关键词:pulmonary anatomical structure;pulmonary computed tomography image;dataset;segmentation of images;medical imaging383|647|0更新时间:2024-05-07
摘要:ObjectiveImages-based segmentation of pulmonary anatomy has been set up the anatomical structures to formulate rapid and targeted diagnostic information. The purpose of pulmonary anatomy segmentation has been associated to a pixel in an image with an anatomical structure without the need for manual initialization. A lots of supervised deep learning image segmentation have been illustrated for segmenting regions of interest in pulmonary CT(computer tomography) images. The medical image segmentation has greatly relied on high-quality labeled medical image data, CT images-based lung anatomy labeled data has been insufficient adopted due to the lack of expert annotation of regions of interest and the lack of infrastructure and standards for sharing labeled data. Most of pulmonary CT annotation datasets have focused on thoracic cancer, pulmonary nodules, tuberculosis, pneumonia and lung segmentation. A dataset of pulmonary CT/CTA(computer tomography/computer tomography angiography) scan images with labels has facilitated the evolvement of pulmonary anatomical structure segmentation algorithms. The dataset has been evaluated the performance of state-of-the-art pulmonary anatomy structure segmentation methods for chest CT scans. It has been difficult to compare various algorithms for pulmonary anatomy structure segmentation. Different methods have been evaluated on different datasets using different evaluation measures in common. The related dataset has implemented a dataset of chest CT scans to identify varying abnormalities based on the reference standards in the context of airway, lung parenchyma, lobe and pulmonary artery. The vein segmentations have been established. The dataset has a unique calculation to compare pulmonary anatomy structure algorithms via the comparison all methods against the reference standard baseline.MethodA sum of 67 sets of CT/CTA images of the pulmonary have labeled in this dataset including 24 sets of CT images and 43 sets of CTA images via a total of 26 157 slices images. Each set of CT/CTA images have labeled for airway, lung parenchyma, lobe, pulmonary artery and vein. Multi-channel images have represented a variety of regular-based clinical scanners based on a reconstructed mediastinal window algorithm. The medical image-based dataset has been annotated and verified via. Manual corrections have annotated using internal software funded by the Key Laboratory of Medical Imaging Intelligent Computing, Ministry of Education.ResultPart of dataset representative segmentation tasks have been used via pulmonary CT anatomical structure segmentation (conference details: the medical image challenge competition held during the 4th International Symposium on Image Computing and Digital Medicine (ISICDM) in Shenyang, China). The representative dataset has included 10 groups of CT and CTA in the training dataset and 5 groups of test dataset. The challenge competition has also offered a platform for evaluating model performance of pulmonary blood vessels, airways and lung parenchyma. The result of segmentation and the effect of 3D reconstruction have evaluated by Dice coefficients, over-segmentation rate, under-segmentation rate and medical and algorithmic industry experts.ConclusionFour parts of labeled image datasets have been used as a pulmonary CT dataset. This dataset has labeled using different colored pixels and saved respectively for different pulmonary anatomies structure. The annotated data have been re-formatted to ensure easy access. The location of the markers in color pixels has been displayed via 25 000 labeled sliced images dataset using the image format of the raw data. All annotated images from the digital imaging and communications in medicine (DICOM) format to portable network graphics (PNG) images has been converted based on standard DICOM data. The chest CT image dataset has provided valid annotated data via DICOM-based sensitive information re-movement. First each set of CT/CTA has labeled with 4 different target region categories in the context of airway, lung parenchyma, lobe and pulmonary artery and vein to complement the anatomical structure of CT/CTA image dataset of the pulmonary. Next, a partially representative dataset has been and the have verified by the challenge competition. Lastly, clear and intuitive 3-dimensional visualized structural images have been reconstructed for the acquisition of each anatomical structure of the pulmonary segmented via CT/CTA images to assist in the diagnosis of pulmonary diseases. First, this dataset has not annotated lung segments. It has been difficult to obtain that the invisibility of lung segment boundaries based on targeted and accurate reference segmentation criteria. Second, the annotation data have been basically carried out on healthy images and rarely on lesion images. The most important feature of medical datasets has upgraded the diversity of data. The robustness of image segmentation have been implementing further. Last, manual annotation of medical anatomical structure images have inevitably resulted some errors. Supplementing lung segments markers and improving the diversity of data based on pulmonary CT anatomical structure segmentation algorithms have been implementing further via labeling more lesion images.关键词:pulmonary anatomical structure;pulmonary computed tomography image;dataset;segmentation of images;medical imaging383|647|0更新时间:2024-05-07
Dataset
-
 摘要:ObjectiveLiver fibrosis is a common manifestation of many chronic liver diseases. It can develop into cirrhosis and even lead to liver cancer if not treated in time. The early diagnosis of liver fibrosis helps prevent the occurrence of severe liver disease. Studies have shown that timely and correct treatment can reverse liver fibrosis and even cirrhosis. Therefore, the accurate assessment of liver fibrosis is essential to the clinical treatment and prognosis assessment of liver fibrosis. At present, the diagnosis of liver fibrosis in the medical field is evaluated through liver biopsy, which is generally a safe procedure but invasive. The complications of liver biopsy are rare but potentially lethal, so noninvasive diagnosis methods based on imaging have attracted considerable interest.MethodThis paper proposes a network for the segmentation of liver fibrosis regions, called LFSCA-UNet(liver fibrosis region segmentation network based on spatial and channel attention mechanisms-UNet). It has improved the U-Net with two different attention mechanisms. U-Net is a convolutional neural network used for image semantic segmentation. Attention U-Net is an improved version of U-Net, it adds a group of attention gate modules into each skip connection of the original U-Net. The attention gate modules in attention U-Net is a spatial attention mechanism. LFSCA-UNet adds a channel attention mechanism to each skip connection structure. In this study, the efficient channel attention(ECA), which is a channel attention mechanism based on the squeeze and excitation network, was used in implementing the added mechanism. The core idea of the squeeze and excitation network is to allow networks to automatically learn dependencies between channels. This network changes a conventional convolution layer to a convolution layer with a squeeze and excitation block, which can be divided into two parts: squeeze and excitation. The squeezing part uses global pooling to obtain a feature vector of a current convolutional layer feature map, whereas the excitation part uses two fully connected layers with different numbers. The first drop of the dimension and the second upgraded, and finally, the weight of each channel is obtained after sigmoid activation, which is multiplied by the original feature map as the input of the subsequent layer of the network. The efficient channel attention block is an improvement of the squeeze and excitation block, which removes the part of reducing dimension and uses 1 d convolution instead of the fully connected layer. It has better performance and fewer parameters. The CT(computed tomography) images used in this study was obtained from 88 patients with liver fibrosis and provided by the Department of Liver Surgery, Renji Hospital, Shanghai Jiao Tong University School of Medicine. One Nvidia Tesla P100 graphics cards with 16 GB memory were used in training networks, and Python 3.8.5 and PyTorch 1.7.1 were used.ResultThis paper horizontally compared five different experimental networks according to five different indicators, namely, Dice coefficient, Jaccard index, precision, recall (sensitivity), and specificity. LFSCA-UNet gets the highest result of mean Dice coefficient (0.933 3), better than the original U-Net (0.539 6%).ConclusionThis paper verifies that the combination of spatial attention and channel attention mechanisms can effectively improve the segmentation result of liver fibrosis. For the spatial attention module, using the channel attention module in optimizing inputs can increase network stability and optimizing outputs can improve the overall effect of the network.关键词:liver fibrosis;image segmentation;spatial attention mechanism;channel attention mechanism;U-Net160|192|3更新时间:2024-05-07
摘要:ObjectiveLiver fibrosis is a common manifestation of many chronic liver diseases. It can develop into cirrhosis and even lead to liver cancer if not treated in time. The early diagnosis of liver fibrosis helps prevent the occurrence of severe liver disease. Studies have shown that timely and correct treatment can reverse liver fibrosis and even cirrhosis. Therefore, the accurate assessment of liver fibrosis is essential to the clinical treatment and prognosis assessment of liver fibrosis. At present, the diagnosis of liver fibrosis in the medical field is evaluated through liver biopsy, which is generally a safe procedure but invasive. The complications of liver biopsy are rare but potentially lethal, so noninvasive diagnosis methods based on imaging have attracted considerable interest.MethodThis paper proposes a network for the segmentation of liver fibrosis regions, called LFSCA-UNet(liver fibrosis region segmentation network based on spatial and channel attention mechanisms-UNet). It has improved the U-Net with two different attention mechanisms. U-Net is a convolutional neural network used for image semantic segmentation. Attention U-Net is an improved version of U-Net, it adds a group of attention gate modules into each skip connection of the original U-Net. The attention gate modules in attention U-Net is a spatial attention mechanism. LFSCA-UNet adds a channel attention mechanism to each skip connection structure. In this study, the efficient channel attention(ECA), which is a channel attention mechanism based on the squeeze and excitation network, was used in implementing the added mechanism. The core idea of the squeeze and excitation network is to allow networks to automatically learn dependencies between channels. This network changes a conventional convolution layer to a convolution layer with a squeeze and excitation block, which can be divided into two parts: squeeze and excitation. The squeezing part uses global pooling to obtain a feature vector of a current convolutional layer feature map, whereas the excitation part uses two fully connected layers with different numbers. The first drop of the dimension and the second upgraded, and finally, the weight of each channel is obtained after sigmoid activation, which is multiplied by the original feature map as the input of the subsequent layer of the network. The efficient channel attention block is an improvement of the squeeze and excitation block, which removes the part of reducing dimension and uses 1 d convolution instead of the fully connected layer. It has better performance and fewer parameters. The CT(computed tomography) images used in this study was obtained from 88 patients with liver fibrosis and provided by the Department of Liver Surgery, Renji Hospital, Shanghai Jiao Tong University School of Medicine. One Nvidia Tesla P100 graphics cards with 16 GB memory were used in training networks, and Python 3.8.5 and PyTorch 1.7.1 were used.ResultThis paper horizontally compared five different experimental networks according to five different indicators, namely, Dice coefficient, Jaccard index, precision, recall (sensitivity), and specificity. LFSCA-UNet gets the highest result of mean Dice coefficient (0.933 3), better than the original U-Net (0.539 6%).ConclusionThis paper verifies that the combination of spatial attention and channel attention mechanisms can effectively improve the segmentation result of liver fibrosis. For the spatial attention module, using the channel attention module in optimizing inputs can increase network stability and optimizing outputs can improve the overall effect of the network.关键词:liver fibrosis;image segmentation;spatial attention mechanism;channel attention mechanism;U-Net160|192|3更新时间:2024-05-07 -
 摘要:ObjectiveAutomatic segmentation of organs at risk (OAR) in computed tomography (CT) has been an essential part of implementing effective treatment strategies to resist lung and esophageal cancers. Accurate segmentation of organs' tumors can aid to interpretate inherent position and morphological changes for patients via facilitating adaptive and computer assisted radiotherapy. Manual delineation of OAR cannot be customized in the future. Scholors have conducted segmentation manually for heart-based backward esophagus spinal and cord-based upper trachea based on intensity levels and anatomical knowledge Complicated variations in the shape and position of organs and low soft tissue contrast between neighboring organs in CT images have caused emerging errors. The CT-based images for lifting manual segmentation skill for thoracic organs have been caused time-consuming. Nonlinear-based modeling of deep convolutional neural networks (DCNNs) has been presented tremendous capability in medical image segmentation. Multi organ segmentation deep learning skill has been applied in abdominal CT images. The small size and irregular shape for automatic segmentation of the esophagus have not been outreached in comparison with even larger size organs. Two skills have related to 3D medical image segmentation have been implemented via the independent separation of each slice and instant 3D convolution to aggregate information between slices and segment all slices of the CT image in. The single slice segmentation skill cannot be used in the multi-layer dependencies overall. Higher computational cost for slices 3D segmentation has been operated via all layers aggregation. A 2.5D deep learning framework has been illustrated to identify the organs location robustly and refine the boundaries of each organ accurately.MethodThis network segmentation of 2.5D slice sequences under the coronal plane composed of three adjacent slices as input can learn the most distinctive of a single slice deeply. The features have been presented in the connection between slices. The image intensity values of all scans were truncated to the range of[-384, 384] HU to omit the irrelevant information in one step. An emerging attention module called efficient global context has been demonstrated based on the completed U-Net neural network structure. The integration for effective channel attention and global context module have been achieved. The global context information has been demonstrated via calculating the response at a location as the weighted sum of the features of all locations in the input feature map. A model has been built up to identify the correlation for channels. The effective feature map can obtain useful information. The useless information can be deducted. The single view long distance dependency between slice sequences can be captured. Attention has been divided into three modules on the aspect of context modeling module, feature conversion module and fusion module. Unlike the traditional global context module, feature conversion module has not required dimensionality to realize the information interaction between channels. The channel attention can be obtained via one dimensional convolution effectively. The capability of pyramid convolution has been used in the encoding layer part. Extracted multi-scale information and expanded receptive field for the convolution layer can be used via dense connection. The pyramid convolution has adapted convolution kernels on different scales and depths. The increased convolution kernels can be used in parallel to process the input and capture different levels of information. Feature transformation has been processed uniformly and individually in multiple parallel branches. The output of each branch has been integrated into the final output. Multi-scale feature extraction based on adjusting the size of the convolution kernel has been achieved than the receptive field resolution down sampling upgration. Multi-layer dense connection has realized feature multiplexing and ensures maximum information transmission. The integration of pyramid convolution and dense connection has obtained a wider range of information and good quality integrated multi-scale images. The backward gradient flow can be smoother than before. An accurate multi-organs segmentation have required local and global information fusion, decoder with each layer of encoders connecting network and the low level details of different levels of feature maps with high level semantics in order to make full use of multi-scale features and enhance the recognition of feature maps. The irregular and closely connected shape of multi-organs in CT images can be calculated at the end. Deep supervision has been added to learn the feature representations of different layers based on the sophisticated feature map aggregation. The boundaries of organs and excessive segmentation deduction in non-organ images and network training can be enhanced effectively. More accurate segmentation results can be produced finally.ResultIn the public dataset of the segmentation of thoracic organs at risk in CT images(SegTHOR) 2019 challenge, the research has been performed CT scans operation on four thoracic organs (i.e., esophagus, heart, trachea and aorta), take Dice similarity coefficient (DSC) and Hausdorff distance (HD) as main criteria, the Dice coefficients of the esophagus, heart, trachea and aorta in the test samples reached 0.855 1, 0.945 7, 0.923 0 and 0.938 3 separately. The HD distances have achieved 0.302 3, 0.180 5, 0.212 2 and 0.191 8 respectively.ConclusionLow level detailed feature maps can capture rich spatial information to highlight the boundaries of organs. High level semantic features have reflected position information and located organs. Multi scale features and global context integration have been the key step to accurate segmentation. The highest average DSC value and HD obtained for heart and Aorta have achieved its high contrast, regular shape, and larger size compared to the other organs. The esophagus had the lowest average DSC and HD values due to its irregularity and low contrast to identify within CT volumes more difficult. The research has achieved a DSC score of 85.5% for the esophagus on test dataset. Experimental results have shown that the proposed method has beneficial for segmenting high risk organs to strengthen radiation therapy planning.关键词:multi-organ segmentation;pseudo three dimension;efficient global context;pyramid convolution;multi-scale features102|165|6更新时间:2024-05-07
摘要:ObjectiveAutomatic segmentation of organs at risk (OAR) in computed tomography (CT) has been an essential part of implementing effective treatment strategies to resist lung and esophageal cancers. Accurate segmentation of organs' tumors can aid to interpretate inherent position and morphological changes for patients via facilitating adaptive and computer assisted radiotherapy. Manual delineation of OAR cannot be customized in the future. Scholors have conducted segmentation manually for heart-based backward esophagus spinal and cord-based upper trachea based on intensity levels and anatomical knowledge Complicated variations in the shape and position of organs and low soft tissue contrast between neighboring organs in CT images have caused emerging errors. The CT-based images for lifting manual segmentation skill for thoracic organs have been caused time-consuming. Nonlinear-based modeling of deep convolutional neural networks (DCNNs) has been presented tremendous capability in medical image segmentation. Multi organ segmentation deep learning skill has been applied in abdominal CT images. The small size and irregular shape for automatic segmentation of the esophagus have not been outreached in comparison with even larger size organs. Two skills have related to 3D medical image segmentation have been implemented via the independent separation of each slice and instant 3D convolution to aggregate information between slices and segment all slices of the CT image in. The single slice segmentation skill cannot be used in the multi-layer dependencies overall. Higher computational cost for slices 3D segmentation has been operated via all layers aggregation. A 2.5D deep learning framework has been illustrated to identify the organs location robustly and refine the boundaries of each organ accurately.MethodThis network segmentation of 2.5D slice sequences under the coronal plane composed of three adjacent slices as input can learn the most distinctive of a single slice deeply. The features have been presented in the connection between slices. The image intensity values of all scans were truncated to the range of[-384, 384] HU to omit the irrelevant information in one step. An emerging attention module called efficient global context has been demonstrated based on the completed U-Net neural network structure. The integration for effective channel attention and global context module have been achieved. The global context information has been demonstrated via calculating the response at a location as the weighted sum of the features of all locations in the input feature map. A model has been built up to identify the correlation for channels. The effective feature map can obtain useful information. The useless information can be deducted. The single view long distance dependency between slice sequences can be captured. Attention has been divided into three modules on the aspect of context modeling module, feature conversion module and fusion module. Unlike the traditional global context module, feature conversion module has not required dimensionality to realize the information interaction between channels. The channel attention can be obtained via one dimensional convolution effectively. The capability of pyramid convolution has been used in the encoding layer part. Extracted multi-scale information and expanded receptive field for the convolution layer can be used via dense connection. The pyramid convolution has adapted convolution kernels on different scales and depths. The increased convolution kernels can be used in parallel to process the input and capture different levels of information. Feature transformation has been processed uniformly and individually in multiple parallel branches. The output of each branch has been integrated into the final output. Multi-scale feature extraction based on adjusting the size of the convolution kernel has been achieved than the receptive field resolution down sampling upgration. Multi-layer dense connection has realized feature multiplexing and ensures maximum information transmission. The integration of pyramid convolution and dense connection has obtained a wider range of information and good quality integrated multi-scale images. The backward gradient flow can be smoother than before. An accurate multi-organs segmentation have required local and global information fusion, decoder with each layer of encoders connecting network and the low level details of different levels of feature maps with high level semantics in order to make full use of multi-scale features and enhance the recognition of feature maps. The irregular and closely connected shape of multi-organs in CT images can be calculated at the end. Deep supervision has been added to learn the feature representations of different layers based on the sophisticated feature map aggregation. The boundaries of organs and excessive segmentation deduction in non-organ images and network training can be enhanced effectively. More accurate segmentation results can be produced finally.ResultIn the public dataset of the segmentation of thoracic organs at risk in CT images(SegTHOR) 2019 challenge, the research has been performed CT scans operation on four thoracic organs (i.e., esophagus, heart, trachea and aorta), take Dice similarity coefficient (DSC) and Hausdorff distance (HD) as main criteria, the Dice coefficients of the esophagus, heart, trachea and aorta in the test samples reached 0.855 1, 0.945 7, 0.923 0 and 0.938 3 separately. The HD distances have achieved 0.302 3, 0.180 5, 0.212 2 and 0.191 8 respectively.ConclusionLow level detailed feature maps can capture rich spatial information to highlight the boundaries of organs. High level semantic features have reflected position information and located organs. Multi scale features and global context integration have been the key step to accurate segmentation. The highest average DSC value and HD obtained for heart and Aorta have achieved its high contrast, regular shape, and larger size compared to the other organs. The esophagus had the lowest average DSC and HD values due to its irregularity and low contrast to identify within CT volumes more difficult. The research has achieved a DSC score of 85.5% for the esophagus on test dataset. Experimental results have shown that the proposed method has beneficial for segmenting high risk organs to strengthen radiation therapy planning.关键词:multi-organ segmentation;pseudo three dimension;efficient global context;pyramid convolution;multi-scale features102|165|6更新时间:2024-05-07 -
 摘要:ObjectiveAs an important criterion for the diagnosis of early-stage lung cancer, chest computed tomography (CT) images-based pulmonary nodules detection have been implemented via location observation, scope and shape of the lesions. The CT image has been analyzed lung organizational structures like the lung parenchyma and the contextual part, such as hydrops, trachea, bronchus, and ribs. CT images-based lung parenchyma has been hard to interpret automatically and precisely. The precise extraction of lung parenchyma has played a vital role in lung-based diseases analyses. Most of lung segmentation have been conducted based on regular image processing algorithms like threshold or morphological operation. The convolutional neural networks (CNNs) have been used in computerized pulmonary disease analysis. CNN-driven lung segmentation algorithms have been adopted in computer-aided diagnosis (CAD). The U-shape structure has been designed for medical image segmentation based on end-to-end fully convolutional network (FCN) structure. The credibility for biomedical image segmentations have been realized based on the encoding and decoding symmetric network structure. A novel convolutional neural network based on U-Net architecture has been illustrated via integrating attention mechanism and dense atrous convolution (DAC).MethodThe network has contained an encoder and a decoder. The encoder has consisted of convolution and down sampling. The deductible spatial dimension of feature maps have been used to learn more semantic information. And the attention mechanism decoder has been implemented for de-convolution and up-sampling to re-configure the spatial dimension of the feature maps. The decoding mode using attention mechanism has been manipulated to make the target area output more effectively. Meanwhile, the algorithm of lung image segmentation has been used to identify the target-oriented neural network's attention using transmitted skip-connection to improve the weight of the salient feature. The feature resolution capability has been enhanced to the requirements for intensive spatial prediction via pooling consecutive operations and convolution striding. The DAC block has been deployed between the encoder and the decoder to extract multi-scale information of the context sufficiently. The advantages of Inception, ResNet and atrous convolution for the block have been inherited to capture multi-sized features consequently. The max-pooling and up-sampling operators have been utilized to reduce and increase the resolution of feature maps intensively based on the classic U-Net framework, which could lead to feature loss and accuracy reduced problems during training. The original max-pooling and up-sampling operators have been replaced via down-sample and up-sample block with inception structure to widen the multi-filters network and avoid feature loss. The Dice coefficient loss function has been used instead of the cross entropy loss to identify the gap between prediction and ground-truth. The deep learning framework Pytorch have been used on a server with two NVIDIA GeForce RTX 2080Ti graphics cards and each GPU has 11 Gigabyte memory. At the experimental stage, the original images have been resized to 256×256 pixels and 80% of these for training besides the test remaining. The proposed model has been trained for 120 epochs. Based on an initial learning rate of 0.000 1, the Adam has been opted as the optimization algorithm.ResultIn order to verify the efficiency of the proposed method, we conduct multi-compatible verifications called FCN-8 s, U-Net, UNet++, ResU-Net and CE-Net (context encoder network) have been conducted. Four segmentation metrics have been adopted to assess the segmentation. These metrics has evolved the Dice similarity coefficient (DSC), the intersection over union (IoU), sensitivity (SE) and accuracy (ACC). The experimental results on the LUNA16 dataset have demonstrated the priorities in terms of all metrics results. The average Dice similarity coefficient has reached 0.985 9, which has 0.443% higher than the segmentation results of the second-performing CE-Net. The model consequence has achieved 0.972 2, 0.993 8, and 0.982 2 each in terms of IoU, ACC and SE. This second qualified segmentation performance has reached: 0.272%, 0.512% and 0.374% each (more better). Compared with other algorithms, the predictable results of modeling has closer to the label made. The adhesive difficulties on the left and right lung cohesion issue have been resolved well.ConclusionAn encoded/decoded structure in novel convolutional neural network has been integrated via attention mechanism and dense atrous convolution for lungs segmentation. The experiment results have illustrated that the qualified and effective framework for segmenting the lung parenchyma area have its own priority.关键词:lung segmentation;convolutional neural networks(CNN);computer-aided diagnosis(CAD);attention mechanism;dense atrous convolution(DAC)88|102|3更新时间:2024-05-07
摘要:ObjectiveAs an important criterion for the diagnosis of early-stage lung cancer, chest computed tomography (CT) images-based pulmonary nodules detection have been implemented via location observation, scope and shape of the lesions. The CT image has been analyzed lung organizational structures like the lung parenchyma and the contextual part, such as hydrops, trachea, bronchus, and ribs. CT images-based lung parenchyma has been hard to interpret automatically and precisely. The precise extraction of lung parenchyma has played a vital role in lung-based diseases analyses. Most of lung segmentation have been conducted based on regular image processing algorithms like threshold or morphological operation. The convolutional neural networks (CNNs) have been used in computerized pulmonary disease analysis. CNN-driven lung segmentation algorithms have been adopted in computer-aided diagnosis (CAD). The U-shape structure has been designed for medical image segmentation based on end-to-end fully convolutional network (FCN) structure. The credibility for biomedical image segmentations have been realized based on the encoding and decoding symmetric network structure. A novel convolutional neural network based on U-Net architecture has been illustrated via integrating attention mechanism and dense atrous convolution (DAC).MethodThe network has contained an encoder and a decoder. The encoder has consisted of convolution and down sampling. The deductible spatial dimension of feature maps have been used to learn more semantic information. And the attention mechanism decoder has been implemented for de-convolution and up-sampling to re-configure the spatial dimension of the feature maps. The decoding mode using attention mechanism has been manipulated to make the target area output more effectively. Meanwhile, the algorithm of lung image segmentation has been used to identify the target-oriented neural network's attention using transmitted skip-connection to improve the weight of the salient feature. The feature resolution capability has been enhanced to the requirements for intensive spatial prediction via pooling consecutive operations and convolution striding. The DAC block has been deployed between the encoder and the decoder to extract multi-scale information of the context sufficiently. The advantages of Inception, ResNet and atrous convolution for the block have been inherited to capture multi-sized features consequently. The max-pooling and up-sampling operators have been utilized to reduce and increase the resolution of feature maps intensively based on the classic U-Net framework, which could lead to feature loss and accuracy reduced problems during training. The original max-pooling and up-sampling operators have been replaced via down-sample and up-sample block with inception structure to widen the multi-filters network and avoid feature loss. The Dice coefficient loss function has been used instead of the cross entropy loss to identify the gap between prediction and ground-truth. The deep learning framework Pytorch have been used on a server with two NVIDIA GeForce RTX 2080Ti graphics cards and each GPU has 11 Gigabyte memory. At the experimental stage, the original images have been resized to 256×256 pixels and 80% of these for training besides the test remaining. The proposed model has been trained for 120 epochs. Based on an initial learning rate of 0.000 1, the Adam has been opted as the optimization algorithm.ResultIn order to verify the efficiency of the proposed method, we conduct multi-compatible verifications called FCN-8 s, U-Net, UNet++, ResU-Net and CE-Net (context encoder network) have been conducted. Four segmentation metrics have been adopted to assess the segmentation. These metrics has evolved the Dice similarity coefficient (DSC), the intersection over union (IoU), sensitivity (SE) and accuracy (ACC). The experimental results on the LUNA16 dataset have demonstrated the priorities in terms of all metrics results. The average Dice similarity coefficient has reached 0.985 9, which has 0.443% higher than the segmentation results of the second-performing CE-Net. The model consequence has achieved 0.972 2, 0.993 8, and 0.982 2 each in terms of IoU, ACC and SE. This second qualified segmentation performance has reached: 0.272%, 0.512% and 0.374% each (more better). Compared with other algorithms, the predictable results of modeling has closer to the label made. The adhesive difficulties on the left and right lung cohesion issue have been resolved well.ConclusionAn encoded/decoded structure in novel convolutional neural network has been integrated via attention mechanism and dense atrous convolution for lungs segmentation. The experiment results have illustrated that the qualified and effective framework for segmenting the lung parenchyma area have its own priority.关键词:lung segmentation;convolutional neural networks(CNN);computer-aided diagnosis(CAD);attention mechanism;dense atrous convolution(DAC)88|102|3更新时间:2024-05-07 -
 摘要:ObjectiveThe different structure of the nodules have not been clear in terms of the lung computer tomography (CT) images interpretation level for lung nodules and surrounding tissues. The detected image resolution has been reduced due to multiple downsampling of the feature extraction network. The feature map of the network top-layer has been predicted to the losses of spatial information and missing nodules. A algorithm called attention_FPN_RPN(attention_feature pyramid networks-region proposal network) for lung nodule detection have been demonstrated via attention mechanism and feature pyramid network.MethodThe detection performance has been improved for an end-to-end lung nodule detection model training via integrating nodule candidate detection and false positive reduction into a model for joint training. At the beginning, the lung parenchyma has been segmented to form the image dataset for training and testing via threshold and morphology. The designated nodule detection model has been based on the region proposal network (RPN) of the faster R-CNN(region convolutional neural network) network. The network model has been illustrated based on the U-Net network structure using low-dose chest CT thin-slice plain scan images as input and outputs candidate nodules position and probability information. Next, the channel-spatial attention mechanism in multi-layers network has been designed in the feature extraction network via the backbone network-ResNet in terms of the semantic and spatial feature compensation mechanism and the amount of extracted feature information. Multi-layers features have been refined to retain feature information each. The integration of lower and higher dimensional features have been spatially interpreted to obtain feature information with more contextual semantics and spatial location information. The semantic information has been qualified via the top-layer of feature map for prediction though the losses of spatial information extracted. The 3D spatial information in CT sequence has not been be fully utilized. The feature pyramid network has been fused high-dimensional feature maps via rich semantic information and low-dimensional feature maps with location information. Multi-scale information network-based detection for insignificant targets like small nodules and proximal vascular nodules has been enhanced. Data enhancement and hard negative data mining has been adopted to optimize the dataset. The sliding window analysis has been used to crop 208×208×208 patch in the preprocessed CT image-based lung parenchymal area. The step size has been consistent with the size of the nodule probability map based on network analysis. Based on convolution calculation, an overlap of 32 pixels amongst the divided blocks have been cropped to eliminate the undesired boundary. At the end, the probability map of the obtained small image block has been refilled into the CT image space to obtain the final probability based on the same size of the graph and the input CT image. Each voxel in the probability graph has illustrated the intended area of lung nodule. For the output result of each small block, the 100 nodules with the highest probability have been retained. Based on summing all patches to obtain candidate nodules, the candidate nodules with high overlap have merged via non-maximum suppression (NMS) method with a threshold of 0.5 for each image. The nodule based on the highest probability has been opted for each group of candidate nodules in the context of a high degree of overlap.Result10-fold cross-validation on the lung nodule analysis 16(LUNA16) dataset has shown that the sensitivity of the proposed model can be reached 97.13% under the average of 25.99 false positives per scan. In the benchmark, the sensitivity has been increased by 2.53%. The false positive average value has been reduced simultaneously to achieve high sensitivity low false positive, the average sensitivity value of the seven false positive rate points has acquired 0.125, 0.25, 0.5, 1, 2, 4, and 8 respectively. The sensitivity of the proposed model has reached 0. 940 and 0.951 respectively under the average of 4, 8 false positives per scan, which is better than mainstream nodule detection methods.ConclusionIn the context of fewer false positives, the demonstrated nodule detection model has improved the detection results of insignificant targets like 3~10 mm small nodules and proximal vessel nodules based on non-false positive reduction measures. A precise detection for tiny lung nodules has prior to the existing pulmonary nodule detection. The setup of an automatic screening system for small lung nodules and a clinical auxiliary diagnosis system for lung nodules have been benefited from the research.关键词:pulmonary nodule detection;attention mechanism;feature pyramid network(FPN);insignificant target detection;10-fold cross-validation82|74|9更新时间:2024-05-07
摘要:ObjectiveThe different structure of the nodules have not been clear in terms of the lung computer tomography (CT) images interpretation level for lung nodules and surrounding tissues. The detected image resolution has been reduced due to multiple downsampling of the feature extraction network. The feature map of the network top-layer has been predicted to the losses of spatial information and missing nodules. A algorithm called attention_FPN_RPN(attention_feature pyramid networks-region proposal network) for lung nodule detection have been demonstrated via attention mechanism and feature pyramid network.MethodThe detection performance has been improved for an end-to-end lung nodule detection model training via integrating nodule candidate detection and false positive reduction into a model for joint training. At the beginning, the lung parenchyma has been segmented to form the image dataset for training and testing via threshold and morphology. The designated nodule detection model has been based on the region proposal network (RPN) of the faster R-CNN(region convolutional neural network) network. The network model has been illustrated based on the U-Net network structure using low-dose chest CT thin-slice plain scan images as input and outputs candidate nodules position and probability information. Next, the channel-spatial attention mechanism in multi-layers network has been designed in the feature extraction network via the backbone network-ResNet in terms of the semantic and spatial feature compensation mechanism and the amount of extracted feature information. Multi-layers features have been refined to retain feature information each. The integration of lower and higher dimensional features have been spatially interpreted to obtain feature information with more contextual semantics and spatial location information. The semantic information has been qualified via the top-layer of feature map for prediction though the losses of spatial information extracted. The 3D spatial information in CT sequence has not been be fully utilized. The feature pyramid network has been fused high-dimensional feature maps via rich semantic information and low-dimensional feature maps with location information. Multi-scale information network-based detection for insignificant targets like small nodules and proximal vascular nodules has been enhanced. Data enhancement and hard negative data mining has been adopted to optimize the dataset. The sliding window analysis has been used to crop 208×208×208 patch in the preprocessed CT image-based lung parenchymal area. The step size has been consistent with the size of the nodule probability map based on network analysis. Based on convolution calculation, an overlap of 32 pixels amongst the divided blocks have been cropped to eliminate the undesired boundary. At the end, the probability map of the obtained small image block has been refilled into the CT image space to obtain the final probability based on the same size of the graph and the input CT image. Each voxel in the probability graph has illustrated the intended area of lung nodule. For the output result of each small block, the 100 nodules with the highest probability have been retained. Based on summing all patches to obtain candidate nodules, the candidate nodules with high overlap have merged via non-maximum suppression (NMS) method with a threshold of 0.5 for each image. The nodule based on the highest probability has been opted for each group of candidate nodules in the context of a high degree of overlap.Result10-fold cross-validation on the lung nodule analysis 16(LUNA16) dataset has shown that the sensitivity of the proposed model can be reached 97.13% under the average of 25.99 false positives per scan. In the benchmark, the sensitivity has been increased by 2.53%. The false positive average value has been reduced simultaneously to achieve high sensitivity low false positive, the average sensitivity value of the seven false positive rate points has acquired 0.125, 0.25, 0.5, 1, 2, 4, and 8 respectively. The sensitivity of the proposed model has reached 0. 940 and 0.951 respectively under the average of 4, 8 false positives per scan, which is better than mainstream nodule detection methods.ConclusionIn the context of fewer false positives, the demonstrated nodule detection model has improved the detection results of insignificant targets like 3~10 mm small nodules and proximal vessel nodules based on non-false positive reduction measures. A precise detection for tiny lung nodules has prior to the existing pulmonary nodule detection. The setup of an automatic screening system for small lung nodules and a clinical auxiliary diagnosis system for lung nodules have been benefited from the research.关键词:pulmonary nodule detection;attention mechanism;feature pyramid network(FPN);insignificant target detection;10-fold cross-validation82|74|9更新时间:2024-05-07 -
 摘要:ObjectiveThe outbreak of corona virus disease 2019 (COVID-19) has become a serious public health event of concern worldwide. The key to controlling the spread of this disease is early detection. Computed tomography (CT) is highly sensitive to the early diagnosis of patients with COVID-19, and the changes in clinical symptoms are time-consistent with the changes in lung CT lesions, which is a simpler, faster indicator for judging changes in the condition. Faint ground-glass opacity is common in the early stage of COVID-19 lesions, and the ground-glass opacity gradually increases as the lesion progresses. Manual detection methods are time consuming, and manual detection inevitably has subjective diagnostic errors. In recent years, deep learning has made great progress in computer vision and achieved outstanding performance in the detection of lung CT scans. In the target detection task, the two-stage target detection method easily achieves a higher precision. The most representative model is faster region convolutional neural network (Faster R-CNN). However, with the increasing diversification and complexity of target detection tasks, the shortcomings of the Faster R-CNN model have also been exposed. In the detection of the ground-glass opacity target, the target size range is large, and Faster R-CNN only uses the highest layer feature map to obtain the region proposal, which has the problem of low recognition rate for small targets. When the region proposal network of Faster R-CNN model supervises the foreground/background classification, most of the overlap calculations between the anchor boxes and the background area are redundant calculations. in the task of detecting ground-glass opacity targets in CT scans of the lung and given the problems of the Faster R-CNN model, an improved method for the feature extraction network and region proposal network of the Faster R-CNN model is proposed.MethodFirst, the feature pyramid network replaces the feature extraction network of Faster R-CNN to generate a feature pyramid. Then, the region proposal network based on location mapping generates anchor boxes and calculates the distance from the center of each anchor boxes to the center of the real object, which is represented by the parameter "centrality". The anchor box judged as the foreground by the region proposal network is further modified as a region proposal, and the foreground/background classification confidence predicted by the region proposal network and centrality are combined as the sorting basis for the region proposal. The interest regions are filtered out from region proposals through non-maximal suppression. Finally, the characteristic regions corresponding to regions of interest are sent to the classification regression network to obtain the detection results. Content of main experiments, the experiment uses recall, mean average precision (mAP), and frames per second (FPS) as evaluation indicators to compare the performance of the standard Faster R-CNN, Faster R-CNN + FPS, and the proposed model, and the effects of different backbone networks on the model in this paper.ResultOn the dataset of COVID-19, the experimental results show that compared with the Faster R-CNN model, the improved model increases recall by 7%, mAP by 3.9%, and FPS from 5 to 9.ConclusionThe improved model can effectively detect the ground-glass opacity target of the patient's lung CT scans and is suitable for small targets. The improved region proposal network reduces network output parameters, saves calculation time, and increases model running speed. Meanings using the feature pyramid network to replace the feature extraction network of Faster R-CNN can be a general method to solve the problem of a large size range of target objects. The method of using the location mapping-based region proposal network to replace the traditional multianchor box mapping-based region proposal network can also provide a reference for accelerating the running speed of the model.关键词:corona virus disease 2019 (COVID-19);ground-glass opacity;faster region convolutional neural network (Faster R-CNN);feature pyramid network (FPN);region proposal network (RPN);residual neural network (ResNet)49|24|2更新时间:2024-05-07
摘要:ObjectiveThe outbreak of corona virus disease 2019 (COVID-19) has become a serious public health event of concern worldwide. The key to controlling the spread of this disease is early detection. Computed tomography (CT) is highly sensitive to the early diagnosis of patients with COVID-19, and the changes in clinical symptoms are time-consistent with the changes in lung CT lesions, which is a simpler, faster indicator for judging changes in the condition. Faint ground-glass opacity is common in the early stage of COVID-19 lesions, and the ground-glass opacity gradually increases as the lesion progresses. Manual detection methods are time consuming, and manual detection inevitably has subjective diagnostic errors. In recent years, deep learning has made great progress in computer vision and achieved outstanding performance in the detection of lung CT scans. In the target detection task, the two-stage target detection method easily achieves a higher precision. The most representative model is faster region convolutional neural network (Faster R-CNN). However, with the increasing diversification and complexity of target detection tasks, the shortcomings of the Faster R-CNN model have also been exposed. In the detection of the ground-glass opacity target, the target size range is large, and Faster R-CNN only uses the highest layer feature map to obtain the region proposal, which has the problem of low recognition rate for small targets. When the region proposal network of Faster R-CNN model supervises the foreground/background classification, most of the overlap calculations between the anchor boxes and the background area are redundant calculations. in the task of detecting ground-glass opacity targets in CT scans of the lung and given the problems of the Faster R-CNN model, an improved method for the feature extraction network and region proposal network of the Faster R-CNN model is proposed.MethodFirst, the feature pyramid network replaces the feature extraction network of Faster R-CNN to generate a feature pyramid. Then, the region proposal network based on location mapping generates anchor boxes and calculates the distance from the center of each anchor boxes to the center of the real object, which is represented by the parameter "centrality". The anchor box judged as the foreground by the region proposal network is further modified as a region proposal, and the foreground/background classification confidence predicted by the region proposal network and centrality are combined as the sorting basis for the region proposal. The interest regions are filtered out from region proposals through non-maximal suppression. Finally, the characteristic regions corresponding to regions of interest are sent to the classification regression network to obtain the detection results. Content of main experiments, the experiment uses recall, mean average precision (mAP), and frames per second (FPS) as evaluation indicators to compare the performance of the standard Faster R-CNN, Faster R-CNN + FPS, and the proposed model, and the effects of different backbone networks on the model in this paper.ResultOn the dataset of COVID-19, the experimental results show that compared with the Faster R-CNN model, the improved model increases recall by 7%, mAP by 3.9%, and FPS from 5 to 9.ConclusionThe improved model can effectively detect the ground-glass opacity target of the patient's lung CT scans and is suitable for small targets. The improved region proposal network reduces network output parameters, saves calculation time, and increases model running speed. Meanings using the feature pyramid network to replace the feature extraction network of Faster R-CNN can be a general method to solve the problem of a large size range of target objects. The method of using the location mapping-based region proposal network to replace the traditional multianchor box mapping-based region proposal network can also provide a reference for accelerating the running speed of the model.关键词:corona virus disease 2019 (COVID-19);ground-glass opacity;faster region convolutional neural network (Faster R-CNN);feature pyramid network (FPN);region proposal network (RPN);residual neural network (ResNet)49|24|2更新时间:2024-05-07 -
 摘要:ObjectiveSince frequent orthopedic diseases cause serious harm to human body, automatic analysis and detection of bone tissue position in computed tomography (CT) has crucial clinical significance for early diagnosis of orthopedic diseases. The method based on manual analysis and diagnosis of bone tissue in CT image has problems such as low efficiency. The accuracy and objective consistency of diagnosis cannot be achieved. Therefore, A cascaded neural network model for bone tissue lesion detection has been demonstrated to aid decision support for orthopedic surgeons' diagnosis.MethodThe proposed bone lesion detection algorithm has mainly consist of four steps. At first, convert original data into CT value data in terms of the conversion formula and relative files that have been illustrated by the National Institutes of Health Clinical Center (NIHCC), USA, in the preprocessing stage. The segmentation has been conducted via the mean value of CT value data as the threshold in order to filter out most of the non-human body parts in the image. The segmentation cannot be filtered out entirely due to the high CT value of the bed plate material of CT equipment. Rectangle kernels (RK)-based opening operation in morphological operations have benefited to filter out the CT bed plate from CT image. According to the characteristics of bone tissue in CT image, a contrast enhancement method based on Gamma transform is proposed to enhance the contrast of CT images. Next, the approximate bone tissue area in enhanced CT images have been calculated via thresholding based on the distribution range of the Hounsfield unit (HU) of the bone tissue in the CT image. The cascaded object detection model has been set up as our baseline. The attention mechanism and deformable convolution have been integrated to increase the global context relevance of the feature map based on the bone lesions with variable shapes. At last, the feature fusion module has been used to strengthen the fusion of feature information at various scales. A multi-scale feature map for the training and prediction of bone tissue lesions has been sorted out.ResultFour designated groups of comparative experiments in the context of the network structure have compared with modeling capability. The detected model has been mainly examined based on average precision (AP). 1) ResNet50, ResNet101 and ResNeXt101 have been used as feature extraction networks to complete training and testing based on naïve Cascade R-CNN(region-convolutional neural network) model to calculate the baseline. The analyzed results have shown that ResNeXt101 has the best optimization based on the AP up to 0.543 to get the baseline. 2) Feature pyramid networks (FPN), path aggregation feature pyramid networks (PAFPN), neural architecture search-feature pyramid networks (NAS-FPN) and naive structure have been adapted to complete model training and testing based on the calculated Cascade R-CNN. The feature fusion module based on the best value has been sorted out. The best PAFPN based on the AP increased to 0.721 has been leaked out the feature fusion module. 3) Two groups of comparative experiments have been illustrated. Firstly, batch normalization (BN) and group normalization (GN) modules have been used in the head of R-CNN for entire training and testing. The results have shown that the performance of GN is better than BN with 0.723 AP. Attention mechanism block and deformable convolution block have been embedded in model to verify their effectiveness in the next step. The verified results have shown the effectiveness of attention mechanism and deformable convolution module. The AP have been improved to 0.816. 4) The trained model and other object detection network models have been calculated to compare the testing value of each model. The research experiments results have been achieved based on DeepLesion dataset. The results have shown as below: 1) the recall is 0.85; 2) the precision is 0.613; 3) the F1-score is 0.712; 4) the AP is 0.816. The performance has been significantly improved in comparison of the existing universal CT lesion detection models based on the recall rates of 0.574 and 0.70.ConclusionThe main methods such as HU value threshold segmentation and morphological operations have been used to filter out most of the non-bone tissue area in the CT image at the image preprocessing stage. The bone tissue area has been highlighted coupled with the enhancement of the image contrast further. The training model has been accelerated to reduce the interference of noise. The second group of experimental results have improved the fusion of low-level feature information, high-level feature information and enhances the location information of high-level features. The semantic information of low-level features based on the multi-scale feature pyramid fusion module has been embedded in the network structure. The detection performance has been significantly improved at the end. The third group of experimental results have been concluded based on the enhancing adaptation of attention mechanism and the weight of irrelevant information deduction. The operation of deformable convolution module has realized the network adaptation further based on multi-shapes and sizes convolution kernels the fourth group of experiment results have achieved via the comparison experiment between our model and other object detection models. The metrics including recall, precision, F1-score and AP have been mainly evaluated in these models. The experimental results have demonstrated that the model analysis has a good detection effect on the CT bone tissue lesions in the context of upgrading diagnostic efficiency, missed diagnosis deduction and quick diagnosis and treatment. The differential diagnosis of bone tissue lesions has been aided effectively. The real-time detection capability can be strengthened via the deduction of model parameters quantity and the time of training and judging.关键词:bone lesion detection;multi-scale object detection;attention mechanism;medical image process;Cascade R-CNN71|161|1更新时间:2024-05-07
摘要:ObjectiveSince frequent orthopedic diseases cause serious harm to human body, automatic analysis and detection of bone tissue position in computed tomography (CT) has crucial clinical significance for early diagnosis of orthopedic diseases. The method based on manual analysis and diagnosis of bone tissue in CT image has problems such as low efficiency. The accuracy and objective consistency of diagnosis cannot be achieved. Therefore, A cascaded neural network model for bone tissue lesion detection has been demonstrated to aid decision support for orthopedic surgeons' diagnosis.MethodThe proposed bone lesion detection algorithm has mainly consist of four steps. At first, convert original data into CT value data in terms of the conversion formula and relative files that have been illustrated by the National Institutes of Health Clinical Center (NIHCC), USA, in the preprocessing stage. The segmentation has been conducted via the mean value of CT value data as the threshold in order to filter out most of the non-human body parts in the image. The segmentation cannot be filtered out entirely due to the high CT value of the bed plate material of CT equipment. Rectangle kernels (RK)-based opening operation in morphological operations have benefited to filter out the CT bed plate from CT image. According to the characteristics of bone tissue in CT image, a contrast enhancement method based on Gamma transform is proposed to enhance the contrast of CT images. Next, the approximate bone tissue area in enhanced CT images have been calculated via thresholding based on the distribution range of the Hounsfield unit (HU) of the bone tissue in the CT image. The cascaded object detection model has been set up as our baseline. The attention mechanism and deformable convolution have been integrated to increase the global context relevance of the feature map based on the bone lesions with variable shapes. At last, the feature fusion module has been used to strengthen the fusion of feature information at various scales. A multi-scale feature map for the training and prediction of bone tissue lesions has been sorted out.ResultFour designated groups of comparative experiments in the context of the network structure have compared with modeling capability. The detected model has been mainly examined based on average precision (AP). 1) ResNet50, ResNet101 and ResNeXt101 have been used as feature extraction networks to complete training and testing based on naïve Cascade R-CNN(region-convolutional neural network) model to calculate the baseline. The analyzed results have shown that ResNeXt101 has the best optimization based on the AP up to 0.543 to get the baseline. 2) Feature pyramid networks (FPN), path aggregation feature pyramid networks (PAFPN), neural architecture search-feature pyramid networks (NAS-FPN) and naive structure have been adapted to complete model training and testing based on the calculated Cascade R-CNN. The feature fusion module based on the best value has been sorted out. The best PAFPN based on the AP increased to 0.721 has been leaked out the feature fusion module. 3) Two groups of comparative experiments have been illustrated. Firstly, batch normalization (BN) and group normalization (GN) modules have been used in the head of R-CNN for entire training and testing. The results have shown that the performance of GN is better than BN with 0.723 AP. Attention mechanism block and deformable convolution block have been embedded in model to verify their effectiveness in the next step. The verified results have shown the effectiveness of attention mechanism and deformable convolution module. The AP have been improved to 0.816. 4) The trained model and other object detection network models have been calculated to compare the testing value of each model. The research experiments results have been achieved based on DeepLesion dataset. The results have shown as below: 1) the recall is 0.85; 2) the precision is 0.613; 3) the F1-score is 0.712; 4) the AP is 0.816. The performance has been significantly improved in comparison of the existing universal CT lesion detection models based on the recall rates of 0.574 and 0.70.ConclusionThe main methods such as HU value threshold segmentation and morphological operations have been used to filter out most of the non-bone tissue area in the CT image at the image preprocessing stage. The bone tissue area has been highlighted coupled with the enhancement of the image contrast further. The training model has been accelerated to reduce the interference of noise. The second group of experimental results have improved the fusion of low-level feature information, high-level feature information and enhances the location information of high-level features. The semantic information of low-level features based on the multi-scale feature pyramid fusion module has been embedded in the network structure. The detection performance has been significantly improved at the end. The third group of experimental results have been concluded based on the enhancing adaptation of attention mechanism and the weight of irrelevant information deduction. The operation of deformable convolution module has realized the network adaptation further based on multi-shapes and sizes convolution kernels the fourth group of experiment results have achieved via the comparison experiment between our model and other object detection models. The metrics including recall, precision, F1-score and AP have been mainly evaluated in these models. The experimental results have demonstrated that the model analysis has a good detection effect on the CT bone tissue lesions in the context of upgrading diagnostic efficiency, missed diagnosis deduction and quick diagnosis and treatment. The differential diagnosis of bone tissue lesions has been aided effectively. The real-time detection capability can be strengthened via the deduction of model parameters quantity and the time of training and judging.关键词:bone lesion detection;multi-scale object detection;attention mechanism;medical image process;Cascade R-CNN71|161|1更新时间:2024-05-07
Computerd Tomography Image
-
 摘要:ObjectiveAccurate diagnosis and early prognosis of breast cancer can increase the survival rates of breast cancer patients. In clinical applications, the process of breast cancer treatment often contains neoadjuvant chemotherapy (NAC) which attempts to reduce tumor size and increase the chance of breast-conserving surgery. However, some patients do not respond positively to NAC and do not show a pathologically complete response. For these patients, NAC is time consuming and highly risky. Therefore, exploring an efficient method for precisely predicting NAC response is essential. A potential scheme is to use medical imaging techniques, such as magnetic resonance imaging in building a computer-assisted diagnosis (CAD) system for predicting NAC response. Most existing CAD methods focus on tumor features, which are highly related to region of interest (ROI) segmentations. At present, breast tumor is segmented manually, and this method cannot satisfy real-time and accurate segmentation requirements. Automatic breast tumor segmentation is a potential way to deal with such issue. Although numerous works about breast tumor segmentation have been proposed and some of them have achieved good results, they mainly focus on the segmentation of single-center datasets. How to improve the generalization ability of a model and ensure its good performance in multicenter datasets is still presents great challenge. To address this problem, we proposed a semantic Laplacian pyramid network (SLAPNet) for segmenting breast tumor with multicenter datasets.MethodSLAPNet is composed of Gaussian and semantic pyramids. The Gaussian pyramid is used for creating multilevel inputs to enable the model to notice not only global image features, such as shape and gray-level distribution, but also local image features, such as edges and textures. It is implemented by smoothing and downsampling input images with Gaussian filters, which can denoise the images and blur details. Thus, the characteristics of large structures in the images can be highlighted. By combining these multiscale inputs, SLAPNet is more robust and generalized, so it can handle irregular objects. The semantic pyramid is produced first after UNet extracts deep semantic features with multilevel inputs and then connects adjacent layers to transfer deep semantic features to different layers. This strategy fuses multi-semantic-level and multilevel features to improve model performance. To reduce the influence of class imbalance, we selected Dice loss as our loss function. To validate the superiority of the proposed method, we trained SLAPNet and other state-of-the-art models with multicenter datasets. Finally, the accuracy (ACC), specificity, sensitivity (SEN), Dice similarity coefficient (DSC), precision, and Jaccard coefficient were used in quantitatively analyzing the segmentation results.ResultCompared with Attention UNet, DeeplabV3, fully convolutional network(FCN), pyramid scene parsing network(PSPNet), UNet, UNet3+, multiscale dual attention network(MSDNet), and pyramid convolutional network(PyConvUNet), the DSC of our model was the highest, with a value of 0.83 when the model was tested on the dataset acquired from Harbin Medical University Cancer Hospital and a value of 0.77 when the model was tested on the public I-SPY 1(investigation of aerial studies to predict your therapeutic response with imaging and moLecular analysis 1) dataset, increasing by at least 1.3%. The visualization results illustrated that SLAPNet produced a small amount of misclassification and omission in the marginal regions and the segmented edge was better than the segmented edges of the other models. The visualization results of error maps indicated that SLAPNet outperformed other models in breast tumor segmentation. Finally, to further validate the stability of the proposed model, we provided the boxplots of the evaluation metrics, which demonstrated that the DSC, Jaccard coefficient, SEN, and ACC of the proposed model were higher than those of the other models and the three quartiles of the proposed model were closer, indicating that SLAPNet was more stable for multicenter breast tumor segmentation.ConclusionThe semantic Laplacian pyramid network proposed in this paper extracted deep semantic features from multilevel inputs and then fused multiscale semantic deep features. This structure guaranteed the high expressive ability of the deep features. We were able to capture more expressive features related to image details by combining multiscale semantic features. Therefore, our proposed model can better distinguish edges and texture features in tumors. The results demonstrated that the pyramid model showed the best performance in multicenter breast cancer tumor segmentation.关键词:breast tumor segmentation;deep learning;semantic pyramids;multiscale semantic feature;multicenter dataset79|122|1更新时间:2024-05-07
摘要:ObjectiveAccurate diagnosis and early prognosis of breast cancer can increase the survival rates of breast cancer patients. In clinical applications, the process of breast cancer treatment often contains neoadjuvant chemotherapy (NAC) which attempts to reduce tumor size and increase the chance of breast-conserving surgery. However, some patients do not respond positively to NAC and do not show a pathologically complete response. For these patients, NAC is time consuming and highly risky. Therefore, exploring an efficient method for precisely predicting NAC response is essential. A potential scheme is to use medical imaging techniques, such as magnetic resonance imaging in building a computer-assisted diagnosis (CAD) system for predicting NAC response. Most existing CAD methods focus on tumor features, which are highly related to region of interest (ROI) segmentations. At present, breast tumor is segmented manually, and this method cannot satisfy real-time and accurate segmentation requirements. Automatic breast tumor segmentation is a potential way to deal with such issue. Although numerous works about breast tumor segmentation have been proposed and some of them have achieved good results, they mainly focus on the segmentation of single-center datasets. How to improve the generalization ability of a model and ensure its good performance in multicenter datasets is still presents great challenge. To address this problem, we proposed a semantic Laplacian pyramid network (SLAPNet) for segmenting breast tumor with multicenter datasets.MethodSLAPNet is composed of Gaussian and semantic pyramids. The Gaussian pyramid is used for creating multilevel inputs to enable the model to notice not only global image features, such as shape and gray-level distribution, but also local image features, such as edges and textures. It is implemented by smoothing and downsampling input images with Gaussian filters, which can denoise the images and blur details. Thus, the characteristics of large structures in the images can be highlighted. By combining these multiscale inputs, SLAPNet is more robust and generalized, so it can handle irregular objects. The semantic pyramid is produced first after UNet extracts deep semantic features with multilevel inputs and then connects adjacent layers to transfer deep semantic features to different layers. This strategy fuses multi-semantic-level and multilevel features to improve model performance. To reduce the influence of class imbalance, we selected Dice loss as our loss function. To validate the superiority of the proposed method, we trained SLAPNet and other state-of-the-art models with multicenter datasets. Finally, the accuracy (ACC), specificity, sensitivity (SEN), Dice similarity coefficient (DSC), precision, and Jaccard coefficient were used in quantitatively analyzing the segmentation results.ResultCompared with Attention UNet, DeeplabV3, fully convolutional network(FCN), pyramid scene parsing network(PSPNet), UNet, UNet3+, multiscale dual attention network(MSDNet), and pyramid convolutional network(PyConvUNet), the DSC of our model was the highest, with a value of 0.83 when the model was tested on the dataset acquired from Harbin Medical University Cancer Hospital and a value of 0.77 when the model was tested on the public I-SPY 1(investigation of aerial studies to predict your therapeutic response with imaging and moLecular analysis 1) dataset, increasing by at least 1.3%. The visualization results illustrated that SLAPNet produced a small amount of misclassification and omission in the marginal regions and the segmented edge was better than the segmented edges of the other models. The visualization results of error maps indicated that SLAPNet outperformed other models in breast tumor segmentation. Finally, to further validate the stability of the proposed model, we provided the boxplots of the evaluation metrics, which demonstrated that the DSC, Jaccard coefficient, SEN, and ACC of the proposed model were higher than those of the other models and the three quartiles of the proposed model were closer, indicating that SLAPNet was more stable for multicenter breast tumor segmentation.ConclusionThe semantic Laplacian pyramid network proposed in this paper extracted deep semantic features from multilevel inputs and then fused multiscale semantic deep features. This structure guaranteed the high expressive ability of the deep features. We were able to capture more expressive features related to image details by combining multiscale semantic features. Therefore, our proposed model can better distinguish edges and texture features in tumors. The results demonstrated that the pyramid model showed the best performance in multicenter breast cancer tumor segmentation.关键词:breast tumor segmentation;deep learning;semantic pyramids;multiscale semantic feature;multicenter dataset79|122|1更新时间:2024-05-07 -
 摘要:ObjectiveMagnetic resonance (MR) image segmentation of brain tumors is crucial for patient evaluation and treatment. In recent years, as the feature extraction capability of convolutional neural networks improved, deep learning technology has been applied to medical image segmentation and achieved better results than those of traditional segmentation methods. The accuracy of image semantic segmentation depends on the effect of semantic feature extraction and processing. Traditional U-Net integrates high-level and low-level features inefficiently, leading to a loss of effective image information. In addition, U-Net does not make full use of context information. This study proposes a segmentation method for brain tumor MR images based on the attention mechanism and multi-view fusion U-Net algorithm.MethodThe model is modified as follows. First, in order to improve the network structure, residual structure is added to the U-Net to enhance the compensation of the network low-level information to the high-level information. On the one hand, the application of the residual idea, removes the same main part of the output of each convolution layer, highlights the small changes, makes each layer of the network more sensitive to the changes of the output, and makes the training easier, on the other hand, it alleviates the problem of gradient disappearance. Second, attention mechanism is added to the cascade structure, and the weight of tumor region is increased adaptively to enhance the utilization of effective information and improve the segmentation accuracy. Third, multi-scale feature fusion module is used to replace the traditional convolution layer between the down sampling structure and up sampling structure, and hole convolution with different sampling rates is used to extract and fuse the multi-scale feature information of the image in parallel. The above is the adjustment of the network structure, and the influence of loss function and 3D structure of Brain tumor MR data set on model training is also considered in the experiment. There is a class imbalance problem in brain tumor segmentation, tumor area is smaller than normal brain tissue, network training is easily guided by a large number of irrelevant pixels, and linear combination of generalized dice loss and cross entropy loss is used to solve class imbalance problem and accelerate convergence. At the same time, Considering the influence of different view slices on the segmentation performance of the model, the three view slices of 3D MR image are trained respectively, and the multi view training model is fused in the segmentation prediction to improve the segmentation performance of the model.ResultThe proposed model is validated using the brain tumor MR image data set provided by Multi- modal Brain Tumor Segmentation Challenge 2018 (BraTS18), which includes four kinds of MR images of 210 high-grade gliomas(HGG) patients and 75 low-grade gliomas(LGG) patients and their real segmentation labels. The Dice similarity coefficient and Hausdorff distance95 are selected as technical indicators to further evaluate the accuracy of the brain tumor segmentation results. The effectiveness of the proposed module is proved by ablation experiments. After adding all modules, the performance of the model is optimal, the Dice scores of the entire tumor area, the core region, and the enhanced region reach 0. 883, 0. 812, and 0.774. Compared with the traditional U-Net, it is improved by 3.9%, 5.1% and 3.3% respectively, especially in the core region. After the fusion of three perspective slice training, comprehensive experiments show that the Dice scores of the entire tumor area, the core region, and the enhanced region reach 0. 907, 0. 838, and 0. 819 respectively, this algorithm exhibits better performance in terms of Dice score and Hausdorff 95 distance than others. At the same time, the slices showthe slice shows the segmentation comparison with other classical models such as FCN. In the HGG sample, the proposed method has more delicate segmentation effect in the tumor boundary area. However, the segmentation results of the LGG sample is not as good as the HGG sample, this is due to the small number of LGG training samples, which is only one third of HGG samples, proposed method is still performs better than other methods.ConclusionThe segmentation method of brain tumor MR images proposed in this study improves the short- comings of the traditional U-Net network in extracting and using image semantic features, and introduces attention mechanism and multi-scale feature fusion module. The weighted mixed loss function is used to solve the class imbalance problem in brain tumor segmentation, and considering the spatial characteristics of the dataset, the multi-view model is fused to improve the segmentation performance. Experimental results show that the algorithm has good performance in the segmentation of different tumor regions and has stronger robustness, thus providing a useful reference for the clinical application of brain tumor MR image segmentation.关键词:brain tumor segmentation;convolution neural network(CNN);multi scale feature;attention mechanism;multi-view fusion100|536|8更新时间:2024-05-07
摘要:ObjectiveMagnetic resonance (MR) image segmentation of brain tumors is crucial for patient evaluation and treatment. In recent years, as the feature extraction capability of convolutional neural networks improved, deep learning technology has been applied to medical image segmentation and achieved better results than those of traditional segmentation methods. The accuracy of image semantic segmentation depends on the effect of semantic feature extraction and processing. Traditional U-Net integrates high-level and low-level features inefficiently, leading to a loss of effective image information. In addition, U-Net does not make full use of context information. This study proposes a segmentation method for brain tumor MR images based on the attention mechanism and multi-view fusion U-Net algorithm.MethodThe model is modified as follows. First, in order to improve the network structure, residual structure is added to the U-Net to enhance the compensation of the network low-level information to the high-level information. On the one hand, the application of the residual idea, removes the same main part of the output of each convolution layer, highlights the small changes, makes each layer of the network more sensitive to the changes of the output, and makes the training easier, on the other hand, it alleviates the problem of gradient disappearance. Second, attention mechanism is added to the cascade structure, and the weight of tumor region is increased adaptively to enhance the utilization of effective information and improve the segmentation accuracy. Third, multi-scale feature fusion module is used to replace the traditional convolution layer between the down sampling structure and up sampling structure, and hole convolution with different sampling rates is used to extract and fuse the multi-scale feature information of the image in parallel. The above is the adjustment of the network structure, and the influence of loss function and 3D structure of Brain tumor MR data set on model training is also considered in the experiment. There is a class imbalance problem in brain tumor segmentation, tumor area is smaller than normal brain tissue, network training is easily guided by a large number of irrelevant pixels, and linear combination of generalized dice loss and cross entropy loss is used to solve class imbalance problem and accelerate convergence. At the same time, Considering the influence of different view slices on the segmentation performance of the model, the three view slices of 3D MR image are trained respectively, and the multi view training model is fused in the segmentation prediction to improve the segmentation performance of the model.ResultThe proposed model is validated using the brain tumor MR image data set provided by Multi- modal Brain Tumor Segmentation Challenge 2018 (BraTS18), which includes four kinds of MR images of 210 high-grade gliomas(HGG) patients and 75 low-grade gliomas(LGG) patients and their real segmentation labels. The Dice similarity coefficient and Hausdorff distance95 are selected as technical indicators to further evaluate the accuracy of the brain tumor segmentation results. The effectiveness of the proposed module is proved by ablation experiments. After adding all modules, the performance of the model is optimal, the Dice scores of the entire tumor area, the core region, and the enhanced region reach 0. 883, 0. 812, and 0.774. Compared with the traditional U-Net, it is improved by 3.9%, 5.1% and 3.3% respectively, especially in the core region. After the fusion of three perspective slice training, comprehensive experiments show that the Dice scores of the entire tumor area, the core region, and the enhanced region reach 0. 907, 0. 838, and 0. 819 respectively, this algorithm exhibits better performance in terms of Dice score and Hausdorff 95 distance than others. At the same time, the slices showthe slice shows the segmentation comparison with other classical models such as FCN. In the HGG sample, the proposed method has more delicate segmentation effect in the tumor boundary area. However, the segmentation results of the LGG sample is not as good as the HGG sample, this is due to the small number of LGG training samples, which is only one third of HGG samples, proposed method is still performs better than other methods.ConclusionThe segmentation method of brain tumor MR images proposed in this study improves the short- comings of the traditional U-Net network in extracting and using image semantic features, and introduces attention mechanism and multi-scale feature fusion module. The weighted mixed loss function is used to solve the class imbalance problem in brain tumor segmentation, and considering the spatial characteristics of the dataset, the multi-view model is fused to improve the segmentation performance. Experimental results show that the algorithm has good performance in the segmentation of different tumor regions and has stronger robustness, thus providing a useful reference for the clinical application of brain tumor MR image segmentation.关键词:brain tumor segmentation;convolution neural network(CNN);multi scale feature;attention mechanism;multi-view fusion100|536|8更新时间:2024-05-07 -
 摘要:ObjectiveMedical image registration has been widely used on the aspect of clinical diagnosis, treatment, intraoperative navigation, disease prediction and radiotherapy planning. Non-learning registration algorithms have matured nowadays in common. Non-learning-based registration algorithms have optimized the deformation parameters iteratively to cause poor robustness because of the huge limitations in the computation speed. Various deep convolution neural networks (DCNNs) models have been running in medical image registration due to the powerful feature expression and learning. DCNNs-based image registration has been divided into supervised and unsupervised categories. The supervised-learning-based registration algorithms have intensive data requirements, which require locking the anatomical landmarks to identify the deformation areas, the performance of reliability of the landmarks has been greatly relied on even the supervised-learning based registration algorithm plays well. Real label information still cannot be acquired. Scholars have focused on unsupervised image registration to complete the defects of supervised image registration. To assess the deformation parameters of the image pair directly via appropriate optimization goals and deformation area constraints. It is difficult to design accurate metric to quantify the similarity of image pairs because the low multimodal images (MI)-based demonstration accuracy in the context of the quite differences amongst content, grayscale, texture and others. Unsupervised registration has been opted in appropriate image similarity to optimize targets involving mean square error, correlation coefficient and normalized mutual information. Most of these similarity assessments have been based on global gray scale. The local deformation still cannot be assessed accurately via good quality e registration structure. An integrated ensemble attention-based augmentation and dual similarity guidance registration network(EADSG-RegNet) has upgraded the registration accuracy of T2-weighted magnetic resonance image and T1-weighted magnetic resonance template image.MethodEADSG-RegNet network has been designated to assess the deformation area between the moving and fixed image pairs. The feature extraction, deformation field estimation and resampler have been illustrated in the network mentioned above. A cascade encoder and encoder have been designed to realize the multi-scale feature extraction and deformation area assessment based on U-Net structure modification. An integrated attention augmentation module (IAAM) in the cascade encoder to improve feature extraction capabilities have been demonstrated to improve the accuracy of registration. In a word, the extracted features have been learned to decode the deformation area accurately. Integrated attention augmentation module has been applied to generate the weights of feature channels of the global average feature via global average pooling of the input feature map. The global feature channels (the number of channels is
摘要:ObjectiveMedical image registration has been widely used on the aspect of clinical diagnosis, treatment, intraoperative navigation, disease prediction and radiotherapy planning. Non-learning registration algorithms have matured nowadays in common. Non-learning-based registration algorithms have optimized the deformation parameters iteratively to cause poor robustness because of the huge limitations in the computation speed. Various deep convolution neural networks (DCNNs) models have been running in medical image registration due to the powerful feature expression and learning. DCNNs-based image registration has been divided into supervised and unsupervised categories. The supervised-learning-based registration algorithms have intensive data requirements, which require locking the anatomical landmarks to identify the deformation areas, the performance of reliability of the landmarks has been greatly relied on even the supervised-learning based registration algorithm plays well. Real label information still cannot be acquired. Scholars have focused on unsupervised image registration to complete the defects of supervised image registration. To assess the deformation parameters of the image pair directly via appropriate optimization goals and deformation area constraints. It is difficult to design accurate metric to quantify the similarity of image pairs because the low multimodal images (MI)-based demonstration accuracy in the context of the quite differences amongst content, grayscale, texture and others. Unsupervised registration has been opted in appropriate image similarity to optimize targets involving mean square error, correlation coefficient and normalized mutual information. Most of these similarity assessments have been based on global gray scale. The local deformation still cannot be assessed accurately via good quality e registration structure. An integrated ensemble attention-based augmentation and dual similarity guidance registration network(EADSG-RegNet) has upgraded the registration accuracy of T2-weighted magnetic resonance image and T1-weighted magnetic resonance template image.MethodEADSG-RegNet network has been designated to assess the deformation area between the moving and fixed image pairs. The feature extraction, deformation field estimation and resampler have been illustrated in the network mentioned above. A cascade encoder and encoder have been designed to realize the multi-scale feature extraction and deformation area assessment based on U-Net structure modification. An integrated attention augmentation module (IAAM) in the cascade encoder to improve feature extraction capabilities have been demonstrated to improve the accuracy of registration. In a word, the extracted features have been learned to decode the deformation area accurately. Integrated attention augmentation module has been applied to generate the weights of feature channels of the global average feature via global average pooling of the input feature map. The global feature channels (the number of channels is$n$ $n$ $n$ 关键词:multimodal registration;deep learning;unsupervised learning;integrated attention augmentation;dual similarity177|501|2更新时间:2024-05-07 -
 摘要:ObjectiveThe brain tumor has been divided into primary and secondary tumors types. Glioma has been divided into lower glial tumors and higher glial tumors. Magnetic resonance imaging(MRI) has been a vital diagnostic tool for brain tumor analysis, detection and surgical planning. Accurate segmentation of brain tumors has been crucial for diagnosis and treatment planning. Manual segmentation has required senior doctors to spend a lot of time to complete nowadays. Automatic brain tumor segmentation has been applied instead manual segmentation further. The intensified profile of the tumor area has overlapped significantly with a healthy portion.MethodThis research has bridged the gap between the efficiency and accuracy of 3D MRI brain tumor segmentation models. A light-weighted rapid semantic segmentation network called LRUNet has been demonstrated. LRUNet has improved the segmentation accuracy and achieved the effect of lightweight, high precision and rapid semantic segmentation in comparison with the existing network. The amount of parameters of these networks has been deleted compared with the algorithms in order to achieve the lightweight effects. At the beginning, the number of channels in the existed 3D-UNet has deducted by four times in each output layer to reduce the number of network parameters dramatically. Next, existed 3D convolution has been excluded and deep separable convolution has been applied to 3D convolution to reduce the number of network parameters on the premise of maintaining accuracy greatly. At last, the convolution-based feature map has not been beneficial to the model entirely. The weight of parameters based on space and channel compression & excitation module has been strengthened to improve the model in the feature map, to reduce the weight of redundant parameters and to improve the performance of the model. Based on 3D-UNet, the number of channels has been reduced 4 times via each convolution. The network becomes more trainable because fewer channels lead to fewer parameters. Three dimensional depth separable convolutions have de-composed the standard convolution into deep convolution and point convolution of 1×1×1. A standard convolutional layer has been integrated to filter and merge into one output. Deep separable convolution has divided the convolution into two layers for filtering and merging each. The effect of this factorization has greatly reduced computation and model size. The application of deep separable convolution has made the network lightweight to realize fast semantic segmentation. The accuracy of the network has not still been improved. The space and channel compression & excitation module have generated a tensor to represent the importance of the feature map in space or channel direction via compressing and exciting the feature map in space or channel direction. The enhancement of important channels or spatial points has been facilitated. The neglect of unimportant channels or spatial points has been weakened. The space and channel compression & excitation module have yielded the network to remain lightweight under no circumstances of increasing the number of arguments. In addition, the accuracy of the network and the training accuracy of the model have been improved simultaneously. First, the tumors contained in the previously given segmentation map have been synthesized to make larger tumor's training area. Second, the best model of intersection over union(IOU) in the validation set has been the optimal parameters. Thirdly, binary cross entropy(BCE) Dice loss has been adopted as the loss function to solve the class imbalance of the foreground and background of the data set itself. Finally, the predicted results have been submitted online to ensure the fairness of the algorithm.ResultThe model has been tested in the Brain Tumor Segmentation Challenge 2018(BraTS 2018) online validation experiment. The average Dice coefficients of tumor segmentation in whole tumor, core tumor and enhanced tumor region have reached 0.893 6, 0.804 6 and 0.787 2 respectively. Compared with 3D-UNet, S3D-UNET, 3D-ESPNET and other algorithms, LRUNet has not only assured the improvement of accuracy, but also greatly reduced the consumption of computational parameters and computational costs in the network.ConclusionA new light-weighted UNet network with only 0.97 MB parameters has been developed to 31 GB floating point operations per second(FLOPs) approximately. The number of parameters has been acquired only 1/16 of the 3D-UNet and the FLOPs have reached 1/52 of the 3D-UNet. The illustrated verification has demonstrated that the great advantages in both performance and number of network parameters have been leaked out based on calculated algorithm (note: the segmentation results have been closest to the true tag). The lightweight and efficient nature of the network has been beneficial to the large-scale 3D medical data sets processing.关键词:3D image processing;fully convolutional network;magnetic resonance imaging(MRI);rapid semantic segmentation;UNet104|90|8更新时间:2024-05-07
摘要:ObjectiveThe brain tumor has been divided into primary and secondary tumors types. Glioma has been divided into lower glial tumors and higher glial tumors. Magnetic resonance imaging(MRI) has been a vital diagnostic tool for brain tumor analysis, detection and surgical planning. Accurate segmentation of brain tumors has been crucial for diagnosis and treatment planning. Manual segmentation has required senior doctors to spend a lot of time to complete nowadays. Automatic brain tumor segmentation has been applied instead manual segmentation further. The intensified profile of the tumor area has overlapped significantly with a healthy portion.MethodThis research has bridged the gap between the efficiency and accuracy of 3D MRI brain tumor segmentation models. A light-weighted rapid semantic segmentation network called LRUNet has been demonstrated. LRUNet has improved the segmentation accuracy and achieved the effect of lightweight, high precision and rapid semantic segmentation in comparison with the existing network. The amount of parameters of these networks has been deleted compared with the algorithms in order to achieve the lightweight effects. At the beginning, the number of channels in the existed 3D-UNet has deducted by four times in each output layer to reduce the number of network parameters dramatically. Next, existed 3D convolution has been excluded and deep separable convolution has been applied to 3D convolution to reduce the number of network parameters on the premise of maintaining accuracy greatly. At last, the convolution-based feature map has not been beneficial to the model entirely. The weight of parameters based on space and channel compression & excitation module has been strengthened to improve the model in the feature map, to reduce the weight of redundant parameters and to improve the performance of the model. Based on 3D-UNet, the number of channels has been reduced 4 times via each convolution. The network becomes more trainable because fewer channels lead to fewer parameters. Three dimensional depth separable convolutions have de-composed the standard convolution into deep convolution and point convolution of 1×1×1. A standard convolutional layer has been integrated to filter and merge into one output. Deep separable convolution has divided the convolution into two layers for filtering and merging each. The effect of this factorization has greatly reduced computation and model size. The application of deep separable convolution has made the network lightweight to realize fast semantic segmentation. The accuracy of the network has not still been improved. The space and channel compression & excitation module have generated a tensor to represent the importance of the feature map in space or channel direction via compressing and exciting the feature map in space or channel direction. The enhancement of important channels or spatial points has been facilitated. The neglect of unimportant channels or spatial points has been weakened. The space and channel compression & excitation module have yielded the network to remain lightweight under no circumstances of increasing the number of arguments. In addition, the accuracy of the network and the training accuracy of the model have been improved simultaneously. First, the tumors contained in the previously given segmentation map have been synthesized to make larger tumor's training area. Second, the best model of intersection over union(IOU) in the validation set has been the optimal parameters. Thirdly, binary cross entropy(BCE) Dice loss has been adopted as the loss function to solve the class imbalance of the foreground and background of the data set itself. Finally, the predicted results have been submitted online to ensure the fairness of the algorithm.ResultThe model has been tested in the Brain Tumor Segmentation Challenge 2018(BraTS 2018) online validation experiment. The average Dice coefficients of tumor segmentation in whole tumor, core tumor and enhanced tumor region have reached 0.893 6, 0.804 6 and 0.787 2 respectively. Compared with 3D-UNet, S3D-UNET, 3D-ESPNET and other algorithms, LRUNet has not only assured the improvement of accuracy, but also greatly reduced the consumption of computational parameters and computational costs in the network.ConclusionA new light-weighted UNet network with only 0.97 MB parameters has been developed to 31 GB floating point operations per second(FLOPs) approximately. The number of parameters has been acquired only 1/16 of the 3D-UNet and the FLOPs have reached 1/52 of the 3D-UNet. The illustrated verification has demonstrated that the great advantages in both performance and number of network parameters have been leaked out based on calculated algorithm (note: the segmentation results have been closest to the true tag). The lightweight and efficient nature of the network has been beneficial to the large-scale 3D medical data sets processing.关键词:3D image processing;fully convolutional network;magnetic resonance imaging(MRI);rapid semantic segmentation;UNet104|90|8更新时间:2024-05-07 -
 摘要:ObjectiveGlioma grading has been a vital research tool for customized treatment in of the glioma. Glioma grading can be an assessment tool for biopsy and histopathological to resolve invasive and time-consuming issues. A non-invasive scheme for grading gliomas precision has played the key role. A reliable non-invasive grading scheme has been implemented for magnetic resonance imaging (MRI)to facilitate computer-assisted diagnosis system(CAD) for glioma grading. The medical image-based grading has been used manual features to implement image-level tumor analysis. Manual feature-based methods have realized higher area under curve (AUC) based on the variation of image intensity and image deformation analyses constrained generalization capability. The emerging deep learning method has projected deep features more semantic and representative compared with the manual features in generalization. The original data has been projected into semantic space to task-level features for data segmentation compared with image-level features. Deep feature-based models have been more qualifier than manual feature-based models on the aspect of classification tasks. Time-consuming and labor-intensive segmentation has been analyzed in lesion regions for tumor. The prediction have been constrained by the tumor segmentation accuracy. A multi-modality fusion network (AMMFNet) has been applied in grading gliomas instead of tumor segmentation.MethodAMMFNet has been an end-to-end multi-scale model to improve glioma grading performance via deep learning-based multi-modal fusion features. The network has contained three components including multi-modal image feature extraction module, adaptive multi-modal and multi-scale features fusion module and classification module respectively. The feature extraction module has been used for deep features extraction based on the images acquired with four different modalities. The width and depth of model has good quality in extracting semantic features. The adaptive multi-modal and multi-scale features fusing module have intended to learn fusion rules via multi-modal and multi-scale deep features. The fusion of the multi-modal features in the same semantic level via high dimensional convolution layers. An adaptive dimension reduction module have adopted to fuse the features in different semantic levels to realize the different shape of feature maps into the same size. Such reduction module has been built up in three-branch structure based on the unique dimension reducing implementation for each branch. Task-level loss and feature-level loss have been used to train the proposed model to upgrade glioma grading accuracy. The task-level loss has been achieved via weighted cross entropy. The feature-level loss has been used to maximize the intra-class feature similarity and inter-class feature discrepancy via the cosine function of two feature vectors. Based on the public Medical Image Computing and Computer Assisted Intervention Society(MICCAI) 2018 challenge dataset to train and test the proposed model, the accuracy(ACC), specificity(SPE), sensitivity(SEN), positive precision value(PPV), negative precision value(NPV), average F1 score and AUC have been prior to analyze the grading validation.ResultThe AUC has been the highest one with a value of 0.965 comparing with visual geometry group 19-layer net(VGG19), ResNet, SENet, SEResNet, InceptionV4 and NASNet. ACC, SPE, PPV, F1-score have been increased by 2.8%, 2.1%, 1.1%, and 3.1% each at most. The tumor region of interest(ROI) modeling input has been trained. The ACC has been increased by 1.2%. Ablation experiments including replacing the deeper convolutional layer with Resblock as well as adding SE block into fusion module have been further validated via customized learning modules. The ACC, SEN, SPE, PPV, NPV, F1 of AMMFNet using SE fusion block have been increased by 0.9%, 0.1%, 2.5%, 1.0%, 0.6%, 1.2% respectively in the context of baseline.ConclusionThe adaptive multi-modal fusion network has been demonstrated via fused multi-modal features and the multi-scale integration of fused deep features. The multi-modal and multi-scale features integration has been illustrated to capture more expressive features related to image details. The demonstration can be prior to locate the tumor even without lesion areas or segmented tumor. An end-to-end model has the good priority in glioma grading.关键词:glioma grading;deep learning;multimodal feature fusion;multiscale deep feature;end-to-end classification model247|90|0更新时间:2024-05-07
摘要:ObjectiveGlioma grading has been a vital research tool for customized treatment in of the glioma. Glioma grading can be an assessment tool for biopsy and histopathological to resolve invasive and time-consuming issues. A non-invasive scheme for grading gliomas precision has played the key role. A reliable non-invasive grading scheme has been implemented for magnetic resonance imaging (MRI)to facilitate computer-assisted diagnosis system(CAD) for glioma grading. The medical image-based grading has been used manual features to implement image-level tumor analysis. Manual feature-based methods have realized higher area under curve (AUC) based on the variation of image intensity and image deformation analyses constrained generalization capability. The emerging deep learning method has projected deep features more semantic and representative compared with the manual features in generalization. The original data has been projected into semantic space to task-level features for data segmentation compared with image-level features. Deep feature-based models have been more qualifier than manual feature-based models on the aspect of classification tasks. Time-consuming and labor-intensive segmentation has been analyzed in lesion regions for tumor. The prediction have been constrained by the tumor segmentation accuracy. A multi-modality fusion network (AMMFNet) has been applied in grading gliomas instead of tumor segmentation.MethodAMMFNet has been an end-to-end multi-scale model to improve glioma grading performance via deep learning-based multi-modal fusion features. The network has contained three components including multi-modal image feature extraction module, adaptive multi-modal and multi-scale features fusion module and classification module respectively. The feature extraction module has been used for deep features extraction based on the images acquired with four different modalities. The width and depth of model has good quality in extracting semantic features. The adaptive multi-modal and multi-scale features fusing module have intended to learn fusion rules via multi-modal and multi-scale deep features. The fusion of the multi-modal features in the same semantic level via high dimensional convolution layers. An adaptive dimension reduction module have adopted to fuse the features in different semantic levels to realize the different shape of feature maps into the same size. Such reduction module has been built up in three-branch structure based on the unique dimension reducing implementation for each branch. Task-level loss and feature-level loss have been used to train the proposed model to upgrade glioma grading accuracy. The task-level loss has been achieved via weighted cross entropy. The feature-level loss has been used to maximize the intra-class feature similarity and inter-class feature discrepancy via the cosine function of two feature vectors. Based on the public Medical Image Computing and Computer Assisted Intervention Society(MICCAI) 2018 challenge dataset to train and test the proposed model, the accuracy(ACC), specificity(SPE), sensitivity(SEN), positive precision value(PPV), negative precision value(NPV), average F1 score and AUC have been prior to analyze the grading validation.ResultThe AUC has been the highest one with a value of 0.965 comparing with visual geometry group 19-layer net(VGG19), ResNet, SENet, SEResNet, InceptionV4 and NASNet. ACC, SPE, PPV, F1-score have been increased by 2.8%, 2.1%, 1.1%, and 3.1% each at most. The tumor region of interest(ROI) modeling input has been trained. The ACC has been increased by 1.2%. Ablation experiments including replacing the deeper convolutional layer with Resblock as well as adding SE block into fusion module have been further validated via customized learning modules. The ACC, SEN, SPE, PPV, NPV, F1 of AMMFNet using SE fusion block have been increased by 0.9%, 0.1%, 2.5%, 1.0%, 0.6%, 1.2% respectively in the context of baseline.ConclusionThe adaptive multi-modal fusion network has been demonstrated via fused multi-modal features and the multi-scale integration of fused deep features. The multi-modal and multi-scale features integration has been illustrated to capture more expressive features related to image details. The demonstration can be prior to locate the tumor even without lesion areas or segmented tumor. An end-to-end model has the good priority in glioma grading.关键词:glioma grading;deep learning;multimodal feature fusion;multiscale deep feature;end-to-end classification model247|90|0更新时间:2024-05-07
Magnetic Resonance Image
-
 摘要:ObjectiveApart from the neuromuscular system, the brain-computer interface (BCI) has provided an alternative way to convey the intention of the brain. For patients who have lost their ability to control their bodies, the BCI technique can recognize the state of the brain and send control order to external assistive devices, assisting patients in their daily lives. For healthy people, the BCI technique can greatly improve multimedia and video game experience and has promising commercial value. Electroencephalogram (EEG) is a flexible and noninvasive brain monitoring method that has been widely used in BCI systems based on motor imagery. The performance of such systems mainly depends on the classification accuracy of motor imagery EEG. In a motor imagery BCI system, EEG signals are recorded when a subject is imaging a specific movement, such as tongue, hand, or foot movement. Motor imagery is classified according to EEG signals. However, owing to long EEG collection time and obvious individual differences, the number of training samples belonging to one subject is small, seriously affecting the performance of convolutional neural network models in EEG recognition tasks. This paper proposes a mirror convolutional neural network (MCNN) that uses ensemble learning and data augmentation methods to improve the recognition accuracy of motor imagery EEG.MethodThe proposed MCNN model can be built on the basis of any motor imagery EEG recognition convolutional neural network (CNN) model. At the training stage, a sufficient number of samples ensure the successful training of an EEG recognition model. For a CNN-based method, training a CNN model needs large number of training samples to have good performance because of the numerous parameters that must be trained. However, the number of training samples in an EEG recognition task is usually small compared with that in a natural image classification task. Therefore, enlarging the number of EEG in a training set is a simple but efficient way to improve the effect of CNN model training. In this paper, we first proposed a mirror EEG construction method to enlarge a training set. A mirror EEG was constructed according to a source EEG by exchanging the left-side and right-side channels of the source EEG. For the left/right hand motor imagery-based EEG, the label of mirror EEG was set opposite to the label of the source EEG because event-related desynchronization and synchronization occur on the contralateral side of the brain. For other types of motor imagery EEG, for instance, feet or tongue motor imagery, the label of the mirror EEG was set in the same manner as the label of the source EEG. At the training stage, the source EEG and mirror EEG constructed on the basis of the source EEG were combined into an enlarged training set to train the source CNN model. This data augmentation method effectively expanded the training samples. At the prediction stage, MCNN improved the EEG recognition performance with the ensemble learning method. Specifically, the trained source CNN model is copied as the mirror CNN model. The source EEG is input into the source CNN model and the mirror EEG was input into the mirror CNN model. The average output category prediction probability of source CNN model and mirror CNN model was the final category prediction probability. In this way, the ensemble learning idea was applied without extra training session.ResultTo verify the effectiveness and universality of the proposed MCNN model, we constructed it according to three different motor imagery recognition CNN models, namely, the EEGNet, Shallow ConvNet, and Deep ConvNet. Experimental verification was performed on the Brain-Computer Interface Competition Ⅳ datasets 2a and 2b. Compared with the results obtained using the original model, the experimental results indicated that the accuracy of the four-category and two-category classification schemes for motor imagery increased by 4.83% and 4.59%, respectively, showing significant improvements. Enhanced performance was observed in different source CNN models, filters, and datasets.ConclusionThe MCNN model was proposed for the EEG recognition of motor imagery BCI. The ideas of ensemble learning and data augmentation were applied to the design of the MCNN. The result showed that the MCNN considerably improved four-category and two-category motor imagery classification performance. Therefore, the proposed MCNN can greatly improve the performance of motor imagery-based BCIs.关键词:brain-computer interface (BCI);motor imagery;mirror convolutional neural network (MCNN);electroencephalogram (EEG);data augmentation;ensemble learning83|218|1更新时间:2024-05-07
摘要:ObjectiveApart from the neuromuscular system, the brain-computer interface (BCI) has provided an alternative way to convey the intention of the brain. For patients who have lost their ability to control their bodies, the BCI technique can recognize the state of the brain and send control order to external assistive devices, assisting patients in their daily lives. For healthy people, the BCI technique can greatly improve multimedia and video game experience and has promising commercial value. Electroencephalogram (EEG) is a flexible and noninvasive brain monitoring method that has been widely used in BCI systems based on motor imagery. The performance of such systems mainly depends on the classification accuracy of motor imagery EEG. In a motor imagery BCI system, EEG signals are recorded when a subject is imaging a specific movement, such as tongue, hand, or foot movement. Motor imagery is classified according to EEG signals. However, owing to long EEG collection time and obvious individual differences, the number of training samples belonging to one subject is small, seriously affecting the performance of convolutional neural network models in EEG recognition tasks. This paper proposes a mirror convolutional neural network (MCNN) that uses ensemble learning and data augmentation methods to improve the recognition accuracy of motor imagery EEG.MethodThe proposed MCNN model can be built on the basis of any motor imagery EEG recognition convolutional neural network (CNN) model. At the training stage, a sufficient number of samples ensure the successful training of an EEG recognition model. For a CNN-based method, training a CNN model needs large number of training samples to have good performance because of the numerous parameters that must be trained. However, the number of training samples in an EEG recognition task is usually small compared with that in a natural image classification task. Therefore, enlarging the number of EEG in a training set is a simple but efficient way to improve the effect of CNN model training. In this paper, we first proposed a mirror EEG construction method to enlarge a training set. A mirror EEG was constructed according to a source EEG by exchanging the left-side and right-side channels of the source EEG. For the left/right hand motor imagery-based EEG, the label of mirror EEG was set opposite to the label of the source EEG because event-related desynchronization and synchronization occur on the contralateral side of the brain. For other types of motor imagery EEG, for instance, feet or tongue motor imagery, the label of the mirror EEG was set in the same manner as the label of the source EEG. At the training stage, the source EEG and mirror EEG constructed on the basis of the source EEG were combined into an enlarged training set to train the source CNN model. This data augmentation method effectively expanded the training samples. At the prediction stage, MCNN improved the EEG recognition performance with the ensemble learning method. Specifically, the trained source CNN model is copied as the mirror CNN model. The source EEG is input into the source CNN model and the mirror EEG was input into the mirror CNN model. The average output category prediction probability of source CNN model and mirror CNN model was the final category prediction probability. In this way, the ensemble learning idea was applied without extra training session.ResultTo verify the effectiveness and universality of the proposed MCNN model, we constructed it according to three different motor imagery recognition CNN models, namely, the EEGNet, Shallow ConvNet, and Deep ConvNet. Experimental verification was performed on the Brain-Computer Interface Competition Ⅳ datasets 2a and 2b. Compared with the results obtained using the original model, the experimental results indicated that the accuracy of the four-category and two-category classification schemes for motor imagery increased by 4.83% and 4.59%, respectively, showing significant improvements. Enhanced performance was observed in different source CNN models, filters, and datasets.ConclusionThe MCNN model was proposed for the EEG recognition of motor imagery BCI. The ideas of ensemble learning and data augmentation were applied to the design of the MCNN. The result showed that the MCNN considerably improved four-category and two-category motor imagery classification performance. Therefore, the proposed MCNN can greatly improve the performance of motor imagery-based BCIs.关键词:brain-computer interface (BCI);motor imagery;mirror convolutional neural network (MCNN);electroencephalogram (EEG);data augmentation;ensemble learning83|218|1更新时间:2024-05-07 -
 摘要:ObjectiveColorectal polyps (CPs) and ulcerative colitis (UC) have been the typical colorectal diseases with high morbidity. Colorectal polyps have become the main factor to trigger colon cancers. UC has been an unidentified colorectal inflammatory disease nowadays. The clinical manifestations illustrate the phenomena in abdominal pain, diarrhea, and bloody stools. UC and CPs regard as one key factor to cause colon cancer. High-intensity endoscopic images analyses have intended physician to fatigued. computer-aided diagnosis(CAD)-based endoscopic colorectal disease image classification system has been the essential demonstration. The bag-of-visual-words (BoVW) model has been qualified in medical image classification for endoscopic lesion detection. Locality constrained linear coding (LLC) has been one good quality coding methodology in the BoVW model due to its coding performance and fast iteration speed. BoVW divide an image into many patches to extract its local features. The local features have become scale-invariant feature transform (SIFT) and histogram of oriented gradient (HOG). These local features have been coded via LLC. Spatial pyramid matching (SPM) and maximum pooling have been used to preserve the spatial information of the image. Some small polyps and unidentified UC have not been leaked out from the intestinal wall in the endoscopic image. The performance of LLC codebook has not been classified the similar endoscope images. To realize category-specific dictionary and shared dictionary learning (CSDL), this research has aimed to endoscopic colorectal image classification via learning a priori weight shared codebook in the LLC. The priori weight shared codebook has qualified good classification results via the endoscopic images classification.MethodLLC-based priori weight shared codebook has been illustrated. The private features have been represented as much as possible. Two control factors in the LLC cost function have been added in. The rearrangement of the columns for the codebook have been arranged via these rarely used atoms called shared atoms in the bottom of the updated codebook in the coding process. Prior knowledge has been demonstrated to have good performance in image processing. The endoscopic images have potential to the probabilistic priors for image process. A priori diagonal weighting matrix has been added to the shared codebook for the better represent of local features when encoding. The weights have derived from the frequency based on the usage of the atoms. First, the local features have been extracted from image patches. The histogram of local color difference (LCDH) features has been identified the information via the relationship of the color distribution distance instead of the SIFT and HOG features due to the rich color information in the endoscopic images. These features have been clustered into the initial codebook via K-means algorithm. The initial codebook has been updated through online dictionary learning algorithm based on the proposed priori weight shared codebook algorithm. All features have been coded based on the updated codebook. The SPM has been used to preserve the spatial information of the endoscopic images. The support vector machines (SVM) classifier has been trained via the codes to identification the lesion images.ResultThis research has integrated a part of the Kvasir dataset and a part of the hospital cooperation dataset to resolve overfitting issue. First, the classification experiments have been performed based on 800 polyp images and 800 normal images. The size
摘要:ObjectiveColorectal polyps (CPs) and ulcerative colitis (UC) have been the typical colorectal diseases with high morbidity. Colorectal polyps have become the main factor to trigger colon cancers. UC has been an unidentified colorectal inflammatory disease nowadays. The clinical manifestations illustrate the phenomena in abdominal pain, diarrhea, and bloody stools. UC and CPs regard as one key factor to cause colon cancer. High-intensity endoscopic images analyses have intended physician to fatigued. computer-aided diagnosis(CAD)-based endoscopic colorectal disease image classification system has been the essential demonstration. The bag-of-visual-words (BoVW) model has been qualified in medical image classification for endoscopic lesion detection. Locality constrained linear coding (LLC) has been one good quality coding methodology in the BoVW model due to its coding performance and fast iteration speed. BoVW divide an image into many patches to extract its local features. The local features have become scale-invariant feature transform (SIFT) and histogram of oriented gradient (HOG). These local features have been coded via LLC. Spatial pyramid matching (SPM) and maximum pooling have been used to preserve the spatial information of the image. Some small polyps and unidentified UC have not been leaked out from the intestinal wall in the endoscopic image. The performance of LLC codebook has not been classified the similar endoscope images. To realize category-specific dictionary and shared dictionary learning (CSDL), this research has aimed to endoscopic colorectal image classification via learning a priori weight shared codebook in the LLC. The priori weight shared codebook has qualified good classification results via the endoscopic images classification.MethodLLC-based priori weight shared codebook has been illustrated. The private features have been represented as much as possible. Two control factors in the LLC cost function have been added in. The rearrangement of the columns for the codebook have been arranged via these rarely used atoms called shared atoms in the bottom of the updated codebook in the coding process. Prior knowledge has been demonstrated to have good performance in image processing. The endoscopic images have potential to the probabilistic priors for image process. A priori diagonal weighting matrix has been added to the shared codebook for the better represent of local features when encoding. The weights have derived from the frequency based on the usage of the atoms. First, the local features have been extracted from image patches. The histogram of local color difference (LCDH) features has been identified the information via the relationship of the color distribution distance instead of the SIFT and HOG features due to the rich color information in the endoscopic images. These features have been clustered into the initial codebook via K-means algorithm. The initial codebook has been updated through online dictionary learning algorithm based on the proposed priori weight shared codebook algorithm. All features have been coded based on the updated codebook. The SPM has been used to preserve the spatial information of the endoscopic images. The support vector machines (SVM) classifier has been trained via the codes to identification the lesion images.ResultThis research has integrated a part of the Kvasir dataset and a part of the hospital cooperation dataset to resolve overfitting issue. First, the classification experiments have been performed based on 800 polyp images and 800 normal images. The size$ K$ $M $ $ K$ $M $ 关键词:computer-aided diagnosis(CAD);locality constrained linear coding(LLC);priori weight shared codebook;colorectal lesion classification;online dictionary learning algorithm43|33|0更新时间:2024-05-07 -
 摘要:ObjectiveAs the gold standard for the diagnosis of gastric cancer, pathological section has been a hotspot nowadays. The degree of precise detection of the lesion area in the section has rather beneficial to in-situ diagnosis and follow-up treatment. The pathologists have missed some subtle changes in cancerous cells in their practice. Automated gastric cancer cells segmentation has been aided to diagnose. Deep learning-based pathological section image of stomach have been obtained qualified classification via deep convolutional neural networks (DCNNs). Focused segmentation for pathological section has been challenged to some issues as below. First, the color and morphology between gastric cancer cells and normal cells to extract deep features. Second, the different magnifications in pathological section images have been hard to segment in different sizes. A semantic segmentation neural network called attention-dilated-efficient U-Net++ (ADEU-Net) has been demonstrated rather than original U-Net to facilitate the precision of gastric cancer cell segmentation.MethodThe illustrated framework is an encoder-decoder networks which can achieve end-to-end training. The capability of the encoder has affected the segmentation accuracy based on the deep features of pathological section images interpretation. The featured extraction part of EfficientNet has been adopted as the encoder via qualified classification. The initialized weights of EfficientNet have been pre-trained on ImageNet and its structure have been divided into five stages for the skipped cohesions. The decoder has been designed in terms of the structure of U-Net++. The encoder and decoder sub-networks have been via the integration of nested, dense skip pathways. An 8 GB GPU model training has been implemented based on the most skip cohesions in U-Net++. The convolution blocks in the decoder has been re-modified to the gradient transfer issues. An additional module called DBlock has been integrated to enhance the feature extraction capability for multi-sized pathological sections. Three multi-layers dilation rates convolution have been cascaded in DBlock to realize the features in receptive fields. The dilation rates of the stacked dilated convolution layers have been calculated to 1, 2, 5 and the receptive field of each layer has been realized 3, 7, 17 each in terms of the structure of hybrid dilated convolution (HDC). The featured maps have been concatenated by channel and fused via a 1×1 convolution layer to realize multi-scale features simultaneously. The attention mechanism has been used to replace the skip connection between the encoder and the decoder to suppress the feature correspondence of the background region effectively as well. The outputs of the encoder and the decoder of the upper layer have been conducted each based on a 1×1 convolution layer. The optional assigned weights to the parameters of the original feature map have been added together to form the attention gate. deep supervised learning can be altered to solve the low speed convergence in the training process.ResultThe experiments on two datasets called SEED and BOT have been conducted to verify the effectiveness of the method obtained from two gastric cancer cell section segmentation competition. The evaluation metrics of the models have Dice coefficient, sensitivity, pixel-wise accuracy and precision. Different segmentation results have also been calculated visually. First the baseline method has been compared to some classical models on SEED dataset in 18.96% accuracy higher than original U-Net and it has been found that the design of feature extraction has been crucial to the segmentation accuracy. Transfer-learning strategies of the encoder have been improved the results greatly. The further ablation experiments have been performed to each added module to confirm the results of segmentation., The Dice coefficient, sensitivity, accuracy and precision has been increased by 5.17% and 0.47%, 2.7% and 0.06%, 3.69% and 4.30%, 4.08% and 6.08% each compared with the baseline model's results with SEED and BOT. The results have demonstrated the effectiveness of each part of proposed algorithm. The visual segmentation results have more similar to the ground truth label.ConclusionA semantic segmentation model called ADEU-Net has been illustrated to the segmentation of pathological sections of gastric cancer task. The involvement of EfficientNet has beneficial to feature extraction, multi-scale features assembling cascade dilated convolution layers and the attention module in replace of the skip connection between the encoder and decoder.关键词:gastric cancer;pathological section;semantic segmentation;deep convolutional neural network(DCNN);attention mechanism;multi-scale features fusion72|172|1更新时间:2024-05-07
摘要:ObjectiveAs the gold standard for the diagnosis of gastric cancer, pathological section has been a hotspot nowadays. The degree of precise detection of the lesion area in the section has rather beneficial to in-situ diagnosis and follow-up treatment. The pathologists have missed some subtle changes in cancerous cells in their practice. Automated gastric cancer cells segmentation has been aided to diagnose. Deep learning-based pathological section image of stomach have been obtained qualified classification via deep convolutional neural networks (DCNNs). Focused segmentation for pathological section has been challenged to some issues as below. First, the color and morphology between gastric cancer cells and normal cells to extract deep features. Second, the different magnifications in pathological section images have been hard to segment in different sizes. A semantic segmentation neural network called attention-dilated-efficient U-Net++ (ADEU-Net) has been demonstrated rather than original U-Net to facilitate the precision of gastric cancer cell segmentation.MethodThe illustrated framework is an encoder-decoder networks which can achieve end-to-end training. The capability of the encoder has affected the segmentation accuracy based on the deep features of pathological section images interpretation. The featured extraction part of EfficientNet has been adopted as the encoder via qualified classification. The initialized weights of EfficientNet have been pre-trained on ImageNet and its structure have been divided into five stages for the skipped cohesions. The decoder has been designed in terms of the structure of U-Net++. The encoder and decoder sub-networks have been via the integration of nested, dense skip pathways. An 8 GB GPU model training has been implemented based on the most skip cohesions in U-Net++. The convolution blocks in the decoder has been re-modified to the gradient transfer issues. An additional module called DBlock has been integrated to enhance the feature extraction capability for multi-sized pathological sections. Three multi-layers dilation rates convolution have been cascaded in DBlock to realize the features in receptive fields. The dilation rates of the stacked dilated convolution layers have been calculated to 1, 2, 5 and the receptive field of each layer has been realized 3, 7, 17 each in terms of the structure of hybrid dilated convolution (HDC). The featured maps have been concatenated by channel and fused via a 1×1 convolution layer to realize multi-scale features simultaneously. The attention mechanism has been used to replace the skip connection between the encoder and the decoder to suppress the feature correspondence of the background region effectively as well. The outputs of the encoder and the decoder of the upper layer have been conducted each based on a 1×1 convolution layer. The optional assigned weights to the parameters of the original feature map have been added together to form the attention gate. deep supervised learning can be altered to solve the low speed convergence in the training process.ResultThe experiments on two datasets called SEED and BOT have been conducted to verify the effectiveness of the method obtained from two gastric cancer cell section segmentation competition. The evaluation metrics of the models have Dice coefficient, sensitivity, pixel-wise accuracy and precision. Different segmentation results have also been calculated visually. First the baseline method has been compared to some classical models on SEED dataset in 18.96% accuracy higher than original U-Net and it has been found that the design of feature extraction has been crucial to the segmentation accuracy. Transfer-learning strategies of the encoder have been improved the results greatly. The further ablation experiments have been performed to each added module to confirm the results of segmentation., The Dice coefficient, sensitivity, accuracy and precision has been increased by 5.17% and 0.47%, 2.7% and 0.06%, 3.69% and 4.30%, 4.08% and 6.08% each compared with the baseline model's results with SEED and BOT. The results have demonstrated the effectiveness of each part of proposed algorithm. The visual segmentation results have more similar to the ground truth label.ConclusionA semantic segmentation model called ADEU-Net has been illustrated to the segmentation of pathological sections of gastric cancer task. The involvement of EfficientNet has beneficial to feature extraction, multi-scale features assembling cascade dilated convolution layers and the attention module in replace of the skip connection between the encoder and decoder.关键词:gastric cancer;pathological section;semantic segmentation;deep convolutional neural network(DCNN);attention mechanism;multi-scale features fusion72|172|1更新时间:2024-05-07 -
 摘要:ObjectiveGlaucoma is a type of disease characterized by atrophy and depression of the nipple, visual field defect, and visual loss. It is one of the main diseases that cause blindness and visual impairment. The clinical diagnosis of glaucoma includes the optic disc, optic cup, intraocular pressure, and the angle between the cornea and iris. Early detection and treatment are crucial for avoiding visual impairment and blindness. In a fundus image, the optic disc is a bright yellow-white spot that resembles a circle, and it is the birthplace of the fundus blood vessels. The shape, size, and depth of the optic disc area and other parameters are important indicators in the clinical diagnosis of glaucoma. Detection of the optic disc can also help determine the location of other lesions, such as fundus hemorrhage and micro aneurysm. Thus, precise detection of the optic disc area is also vital. However, the fundus image is often not bright, and its contrast is poor. In addition, the fundus structure is complex, and the overlapping of tissues and lesions is serious. These problems pose great challenges to the accurate detection and segmentation of the optic disc. The traditional single visual saliency method is considerably affected by color and brightness; hence, tissues, such as blood vessels, and bright lesions easily interfere in optic disc detection. Fully convolutional neural networks (FCNs) classify images at the pixel level, thus solving the problem of image segmentation at the semantic level. In the process of FCN optic disc testing, the detection results are minimally affected by tissues (e.g., vascular lesions), the borders are rough, and the accuracy is low. To overcome these difficulties and achieve accurate detection of the fundus image optic disc, this study combines the advantages of the visual saliency method and FCN, and a visual saliency disc detection method based on the fusion of deep and shallow features is proposed.MethodFirst, on the basis of the prominent characteristics of the optic disc area, a method based on visual saliency detection is used to locate the optic disc area. The saliency detection method based on morphological open reconstruction is utilized to extract the candidate solutions of the optic disc so that the enhanced optic disc becomes conspicuous and appears as a bright circular structure. The center of gravity of the optic disc is obtained and marked, and an image with a pixel size of 400×400 with the center of gravity as the center is extracted to segment the optic disc. Second, the Matconvnet computer vision neural network toolbox is used to extract deep features from the pre-trained model. The pre-trained model uses the pascal-fcn8s-dag.mat file provided on the official website of the Matconvnet toolbox, which uses the PASCAL VOC (pattern analysis, statistical modeling and computational learning visual object classes) 2011 dataset for training. Moreover, the average grayscale of the disc area is calculated for the 400×400 pixel size images to obtain color features. Lastly, by fusing the color feature, depth feature, and background prior information into a single-layer cellular automaton, the similarity is judged based on the distance between different cells in the feature space. Iterative evolution is performed to achieve accurate detection of the optic disc area in the fundus image.ResultThe proposed algorithm is experimentally verified on public datasets of fundus images, namely, DRISHTI-GS, MESSIDOR, and DRIONS-DB. The three datasets are mainly used for optic nerve head detection, including optic cup and optic disc. The optic disc area can be accurately detected in all datasets. The proposed algorithm detects the optic disc area in these datasets, and the evaluation indicators include Dice, Jaccard, recall, and accuracy. In the DRISHTI-GS dataset, Dice is 0.965 8, Jaccard is 0.934 1, recall is 0.964 8, and accuracy is 0.996 6. In the MESSIDOR dataset, Dice, Jaccard, recall, and accuracy have values of 0.961 6, 0.922 4, 0.958 9, and 0.995 3, respectively. In the DRIONS-DB dataset, Dice is 0.971 1, Jaccard is 0.937 6, recall is 0.967 4, and accuracy is 0.996 8. In addition, the proposed algorithm is compared with three algorithms, namely, improved circular Hough transform with Hough peak value selection and red channel superpixel segmentation for optic disc segmentation, accurate optic disc and cup segmentation from fundus images by using a multi-feature-based approach for glaucoma assessment, and dense fully convolutional with U-shape segmentation of the optic disc and cup in color fundus for glaucoma diagnosis. The proposed algorithm has higher accuracy, robustness, and calculation speed than the compared algorithms.ConclusionWe propose a single-layer cellular automaton optic disc detection algorithm that integrates deep and shallow features. Experimental results show that the proposed algorithm can effectively overcome the effects of low brightness and low contrast of fundus images and the interference of blood vessels, lesions, and other tissues. The algorithm is verified on multiple public datasets of fundus images and achieves good detection results. It has strong generalization and realizes accurate detection of the optic disc area.关键词:fundus image;saliency detection;feature fusion;optic disc detection;DRISHTI-GS;MESSIDOR;DRIONS-DB260|182|1更新时间:2024-05-07
摘要:ObjectiveGlaucoma is a type of disease characterized by atrophy and depression of the nipple, visual field defect, and visual loss. It is one of the main diseases that cause blindness and visual impairment. The clinical diagnosis of glaucoma includes the optic disc, optic cup, intraocular pressure, and the angle between the cornea and iris. Early detection and treatment are crucial for avoiding visual impairment and blindness. In a fundus image, the optic disc is a bright yellow-white spot that resembles a circle, and it is the birthplace of the fundus blood vessels. The shape, size, and depth of the optic disc area and other parameters are important indicators in the clinical diagnosis of glaucoma. Detection of the optic disc can also help determine the location of other lesions, such as fundus hemorrhage and micro aneurysm. Thus, precise detection of the optic disc area is also vital. However, the fundus image is often not bright, and its contrast is poor. In addition, the fundus structure is complex, and the overlapping of tissues and lesions is serious. These problems pose great challenges to the accurate detection and segmentation of the optic disc. The traditional single visual saliency method is considerably affected by color and brightness; hence, tissues, such as blood vessels, and bright lesions easily interfere in optic disc detection. Fully convolutional neural networks (FCNs) classify images at the pixel level, thus solving the problem of image segmentation at the semantic level. In the process of FCN optic disc testing, the detection results are minimally affected by tissues (e.g., vascular lesions), the borders are rough, and the accuracy is low. To overcome these difficulties and achieve accurate detection of the fundus image optic disc, this study combines the advantages of the visual saliency method and FCN, and a visual saliency disc detection method based on the fusion of deep and shallow features is proposed.MethodFirst, on the basis of the prominent characteristics of the optic disc area, a method based on visual saliency detection is used to locate the optic disc area. The saliency detection method based on morphological open reconstruction is utilized to extract the candidate solutions of the optic disc so that the enhanced optic disc becomes conspicuous and appears as a bright circular structure. The center of gravity of the optic disc is obtained and marked, and an image with a pixel size of 400×400 with the center of gravity as the center is extracted to segment the optic disc. Second, the Matconvnet computer vision neural network toolbox is used to extract deep features from the pre-trained model. The pre-trained model uses the pascal-fcn8s-dag.mat file provided on the official website of the Matconvnet toolbox, which uses the PASCAL VOC (pattern analysis, statistical modeling and computational learning visual object classes) 2011 dataset for training. Moreover, the average grayscale of the disc area is calculated for the 400×400 pixel size images to obtain color features. Lastly, by fusing the color feature, depth feature, and background prior information into a single-layer cellular automaton, the similarity is judged based on the distance between different cells in the feature space. Iterative evolution is performed to achieve accurate detection of the optic disc area in the fundus image.ResultThe proposed algorithm is experimentally verified on public datasets of fundus images, namely, DRISHTI-GS, MESSIDOR, and DRIONS-DB. The three datasets are mainly used for optic nerve head detection, including optic cup and optic disc. The optic disc area can be accurately detected in all datasets. The proposed algorithm detects the optic disc area in these datasets, and the evaluation indicators include Dice, Jaccard, recall, and accuracy. In the DRISHTI-GS dataset, Dice is 0.965 8, Jaccard is 0.934 1, recall is 0.964 8, and accuracy is 0.996 6. In the MESSIDOR dataset, Dice, Jaccard, recall, and accuracy have values of 0.961 6, 0.922 4, 0.958 9, and 0.995 3, respectively. In the DRIONS-DB dataset, Dice is 0.971 1, Jaccard is 0.937 6, recall is 0.967 4, and accuracy is 0.996 8. In addition, the proposed algorithm is compared with three algorithms, namely, improved circular Hough transform with Hough peak value selection and red channel superpixel segmentation for optic disc segmentation, accurate optic disc and cup segmentation from fundus images by using a multi-feature-based approach for glaucoma assessment, and dense fully convolutional with U-shape segmentation of the optic disc and cup in color fundus for glaucoma diagnosis. The proposed algorithm has higher accuracy, robustness, and calculation speed than the compared algorithms.ConclusionWe propose a single-layer cellular automaton optic disc detection algorithm that integrates deep and shallow features. Experimental results show that the proposed algorithm can effectively overcome the effects of low brightness and low contrast of fundus images and the interference of blood vessels, lesions, and other tissues. The algorithm is verified on multiple public datasets of fundus images and achieves good detection results. It has strong generalization and realizes accurate detection of the optic disc area.关键词:fundus image;saliency detection;feature fusion;optic disc detection;DRISHTI-GS;MESSIDOR;DRIONS-DB260|182|1更新时间:2024-05-07
Research and Application
0

