
最新刊期
卷 25 , 期 10 , 2020
-
 摘要:Deep learning can automatically learn from a large amount of data to obtain effective feature representations, thereby effectively improving the performance of various machine learning tasks. It has been widely used in various fields of medical imaging. Smart healthcare has become an important application area of deep learning, which is an effective approach to solve the following clinical problems: 1) given the limited medical resources, the experienced radiologists are not fully available, which cannot satisfy the fast development of the clinical requirement; 2) the lack of experienced radiologists, which cannot satisfy the fast increase of medical demand. At present, deep learning-based intelligent medical imaging systems are the typical scenarios in smart healthcare. This paper primarily reviews the applications of deep learning methods in various applications using four major clinical imaging techniques (i.e., X-ray, ultrasound, computed tomography(CT), and magnetic resonance imaging(MRI)). These works cover the whole pipeline of medical imaging, including reconstruction, detection, segmentation, registration, and computer-aided diagnosis (CAD). The reviews on medical image reconstruction focus on both MRI reconstruction and low-dose CT reconstruction on the basis of deep learning. Deep learning methods for MRI reconstruction can be divided into two categories: 1) data-driven end-to-end methods, 2) model-based methods. The low-dose CT reconstruction primarily introduces methods on the basis of convolutional neural networks and generative adversarial networks. In addition, deep learning methods for ultrasound imaging, medical image synthesis, and medical image super-resolution are reviewed. The reviews on lesion detection primarily focuses on the deep learning methods for lung lesions detection using CT, the deep learning detection model for tumor lesions, and the deep learning methods for the general lesion area detection. At present, deep learning has been widely used in medical image segmentation tasks, and its performance is significantly improved compared with traditional image segmentation methods. Most deep learning segmentation methods are typical data-driven machine learning models. We review supervised models, semi-supervised models, and self-supervised models with regard to the amount of labeled data and annotation. Medical images contain rich anatomical information, which enhances the performance of deep learning models with different supervision. Deep learning models incorporating prior knowledge are also reviewed. Medical image registration consistency is a difficult task in the field of medical image analysis. Deep learning has become a breakthrough to improve the performance of medical image registration. The end-to-end network structures produce high-precision registration results and have become a hotspot in the field of image registration. Compared with the conventional methods, the deep learning methods for medical image registration have a significant improvement in registration performance. According to the different supervision in the training procedure, this paper divides the deep learning methods for medical image registration into three modes: fully supervised methods, unsupervised methods, and weakly supervised methods. Computer-aided diagnosis is another application of deep learning in the field of medical imaging. This paper summarizes the deep learning methods on CAD with different supervision and the CAD works on the basis of multi-modality medical images. Notably, although deep learning methods have been applied in medical imaging, several challenges are still identified. For example, the small-sample size problem is common in medical imaging analysis. Advanced machine learning methods, including weakly supervised learning, transfer learning, few-shot learning, self-supervised learning, and increase learning, can help alleviate this problem. In addition, the data annotation of medical images is a problem that seriously restricts the extensive and in-depth application of deep learning, and extensive research on automatic data labeling must be carried out. Interpretability of the deep neural networks is also important in medical image analysis. Improving the interpretability of a deep neural network has always been a difficult point, and in-depth research must be carried out in this area. Furthermore, carrying out human-computer collaboration in medical care is important. The lightweight deep neural network is easy to deploy into portable medical devices, giving portable devices more powerful functions, which is also an important research direction. Deep learning has been successful in various tasks in medical imaging analysis. New methods must be developed for its further application in intelligent medical products.关键词:deep learning;medical imaging;image reconstruction;lesion detection;image segmentation;image registration;computer-aided diagnosis(CAD)1473|588|37更新时间:2024-05-07
摘要:Deep learning can automatically learn from a large amount of data to obtain effective feature representations, thereby effectively improving the performance of various machine learning tasks. It has been widely used in various fields of medical imaging. Smart healthcare has become an important application area of deep learning, which is an effective approach to solve the following clinical problems: 1) given the limited medical resources, the experienced radiologists are not fully available, which cannot satisfy the fast development of the clinical requirement; 2) the lack of experienced radiologists, which cannot satisfy the fast increase of medical demand. At present, deep learning-based intelligent medical imaging systems are the typical scenarios in smart healthcare. This paper primarily reviews the applications of deep learning methods in various applications using four major clinical imaging techniques (i.e., X-ray, ultrasound, computed tomography(CT), and magnetic resonance imaging(MRI)). These works cover the whole pipeline of medical imaging, including reconstruction, detection, segmentation, registration, and computer-aided diagnosis (CAD). The reviews on medical image reconstruction focus on both MRI reconstruction and low-dose CT reconstruction on the basis of deep learning. Deep learning methods for MRI reconstruction can be divided into two categories: 1) data-driven end-to-end methods, 2) model-based methods. The low-dose CT reconstruction primarily introduces methods on the basis of convolutional neural networks and generative adversarial networks. In addition, deep learning methods for ultrasound imaging, medical image synthesis, and medical image super-resolution are reviewed. The reviews on lesion detection primarily focuses on the deep learning methods for lung lesions detection using CT, the deep learning detection model for tumor lesions, and the deep learning methods for the general lesion area detection. At present, deep learning has been widely used in medical image segmentation tasks, and its performance is significantly improved compared with traditional image segmentation methods. Most deep learning segmentation methods are typical data-driven machine learning models. We review supervised models, semi-supervised models, and self-supervised models with regard to the amount of labeled data and annotation. Medical images contain rich anatomical information, which enhances the performance of deep learning models with different supervision. Deep learning models incorporating prior knowledge are also reviewed. Medical image registration consistency is a difficult task in the field of medical image analysis. Deep learning has become a breakthrough to improve the performance of medical image registration. The end-to-end network structures produce high-precision registration results and have become a hotspot in the field of image registration. Compared with the conventional methods, the deep learning methods for medical image registration have a significant improvement in registration performance. According to the different supervision in the training procedure, this paper divides the deep learning methods for medical image registration into three modes: fully supervised methods, unsupervised methods, and weakly supervised methods. Computer-aided diagnosis is another application of deep learning in the field of medical imaging. This paper summarizes the deep learning methods on CAD with different supervision and the CAD works on the basis of multi-modality medical images. Notably, although deep learning methods have been applied in medical imaging, several challenges are still identified. For example, the small-sample size problem is common in medical imaging analysis. Advanced machine learning methods, including weakly supervised learning, transfer learning, few-shot learning, self-supervised learning, and increase learning, can help alleviate this problem. In addition, the data annotation of medical images is a problem that seriously restricts the extensive and in-depth application of deep learning, and extensive research on automatic data labeling must be carried out. Interpretability of the deep neural networks is also important in medical image analysis. Improving the interpretability of a deep neural network has always been a difficult point, and in-depth research must be carried out in this area. Furthermore, carrying out human-computer collaboration in medical care is important. The lightweight deep neural network is easy to deploy into portable medical devices, giving portable devices more powerful functions, which is also an important research direction. Deep learning has been successful in various tasks in medical imaging analysis. New methods must be developed for its further application in intelligent medical products.关键词:deep learning;medical imaging;image reconstruction;lesion detection;image segmentation;image registration;computer-aided diagnosis(CAD)1473|588|37更新时间:2024-05-07 -
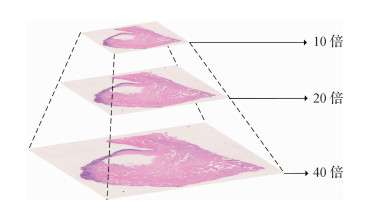 摘要:Histopathology is the gold standard for the clinical diagnosis of tumors and directly related to clinical treatment and prognosis. Its application inclinics has presented challenges in terms of the accuracy and efficiency of histopathological diagnosis, and pathological diagnosis is time consuming and requires pathologists to examine slides with a microscope for them to make reliable decisions. Moreover, the training period of a pathologist is long. In many parts of China, pathology departments are generally overworked due to the insufficient number of pathologists. Recently, deep learning has achieved great success in computer vision. The utilization of whole slide scanners enables the application of deep learning-based classification and segmentation methods to histopathological diagnosis, thereby improving efficiency and accuracy. In this paper, we first introduce the medical background of histopathology diagnosis. Then, we provide an overview of the primary datasets of histopathological diagnosis. We focus on introducing the datasets of three types of malignant tumors along with corresponding computer vision tasks. Breast cancer forms in the cells of breasts and is one of the most common cancer diagnosed in women. Early diagnosis can significantly improve survival rate and quality of life. Sentinel lymph node metastasis are visible when a cancer spreads. The diagnosis of lymph node metastasis is directly related to cancer staging and surgical plan decision. Colon cancer can be detected by colonoscopy biopsy, and early diagnosis requires pathologists to examine slides thoroughly for small malignancies.Computer-aided diagnosis can increase the efficiency of pathologists. Moreover, we propose three key technical problems: data storage and processing, model design and improvement, and learning with small amount or weakly labeled data. Then we review research progress related to tasks, including data preprocessing, classification, and segmentation and transfer learning and multiple instance learning. Pathology datasets are usually stored in a pyramidal tiled image format for fast loading and rescaling. The OpenSlide library provides high-performance pathology data reading, and the open-source softwareautomated slide analysis platform (ASAP) can be used in viewing and labeling these data. Trimming white backgrounds can reduce storage and calculation overhead to 82% on mainstream datasets. A stain normalization technology can eliminate color difference caused by slide production and scanning process. The classification of pathological image patches is the basic structure of whole slide classification and the backbone network of segmentation. Mainstream convolutional neural network models in the field of computer vision, including AlexNet, visual geometry group(VGG), GoogLeNet, and residual neural networks can reach satisfying results for pathological image patches. The patch sampling method can divide a whole slide image into smaller patches that can be processed by the mainstream convolutional neural network models. By aggregating the features of sampled patches through random forest or voting, a patch sampling method can be used in classifying or segmenting arbitrarily sized images. A migration learning technology based on neural network models pretrained on an ImageNet dataset is effective in alleviating the problem introduced by the small number of training samples in histopathological data. Fully convolutional network (FCN) represented by U-Net is a network designed for medical image segmentation tasks and are faster than convolutional neural networks with patch sampling methods. To utilize weakly labeled data, multiple instance learning (MIL) treats whole slide image as a bag of unlabeled pathological image patches. With bags labeled, MIL can be used for weakly supervised learning. Finally, this paper summarizes the main works surveyed and identifies challenges for future research. To make deep learning-based computer aided diagnosis clinically practical, researchers have to improve model accuracy, expand clinical application scenarios, and improve the interpretability of results.关键词:histopathology;deep learning;convolutional neural network;transfer learning;multiple instance learning205|277|9更新时间:2024-05-07
摘要:Histopathology is the gold standard for the clinical diagnosis of tumors and directly related to clinical treatment and prognosis. Its application inclinics has presented challenges in terms of the accuracy and efficiency of histopathological diagnosis, and pathological diagnosis is time consuming and requires pathologists to examine slides with a microscope for them to make reliable decisions. Moreover, the training period of a pathologist is long. In many parts of China, pathology departments are generally overworked due to the insufficient number of pathologists. Recently, deep learning has achieved great success in computer vision. The utilization of whole slide scanners enables the application of deep learning-based classification and segmentation methods to histopathological diagnosis, thereby improving efficiency and accuracy. In this paper, we first introduce the medical background of histopathology diagnosis. Then, we provide an overview of the primary datasets of histopathological diagnosis. We focus on introducing the datasets of three types of malignant tumors along with corresponding computer vision tasks. Breast cancer forms in the cells of breasts and is one of the most common cancer diagnosed in women. Early diagnosis can significantly improve survival rate and quality of life. Sentinel lymph node metastasis are visible when a cancer spreads. The diagnosis of lymph node metastasis is directly related to cancer staging and surgical plan decision. Colon cancer can be detected by colonoscopy biopsy, and early diagnosis requires pathologists to examine slides thoroughly for small malignancies.Computer-aided diagnosis can increase the efficiency of pathologists. Moreover, we propose three key technical problems: data storage and processing, model design and improvement, and learning with small amount or weakly labeled data. Then we review research progress related to tasks, including data preprocessing, classification, and segmentation and transfer learning and multiple instance learning. Pathology datasets are usually stored in a pyramidal tiled image format for fast loading and rescaling. The OpenSlide library provides high-performance pathology data reading, and the open-source softwareautomated slide analysis platform (ASAP) can be used in viewing and labeling these data. Trimming white backgrounds can reduce storage and calculation overhead to 82% on mainstream datasets. A stain normalization technology can eliminate color difference caused by slide production and scanning process. The classification of pathological image patches is the basic structure of whole slide classification and the backbone network of segmentation. Mainstream convolutional neural network models in the field of computer vision, including AlexNet, visual geometry group(VGG), GoogLeNet, and residual neural networks can reach satisfying results for pathological image patches. The patch sampling method can divide a whole slide image into smaller patches that can be processed by the mainstream convolutional neural network models. By aggregating the features of sampled patches through random forest or voting, a patch sampling method can be used in classifying or segmenting arbitrarily sized images. A migration learning technology based on neural network models pretrained on an ImageNet dataset is effective in alleviating the problem introduced by the small number of training samples in histopathological data. Fully convolutional network (FCN) represented by U-Net is a network designed for medical image segmentation tasks and are faster than convolutional neural networks with patch sampling methods. To utilize weakly labeled data, multiple instance learning (MIL) treats whole slide image as a bag of unlabeled pathological image patches. With bags labeled, MIL can be used for weakly supervised learning. Finally, this paper summarizes the main works surveyed and identifies challenges for future research. To make deep learning-based computer aided diagnosis clinically practical, researchers have to improve model accuracy, expand clinical application scenarios, and improve the interpretability of results.关键词:histopathology;deep learning;convolutional neural network;transfer learning;multiple instance learning205|277|9更新时间:2024-05-07 -
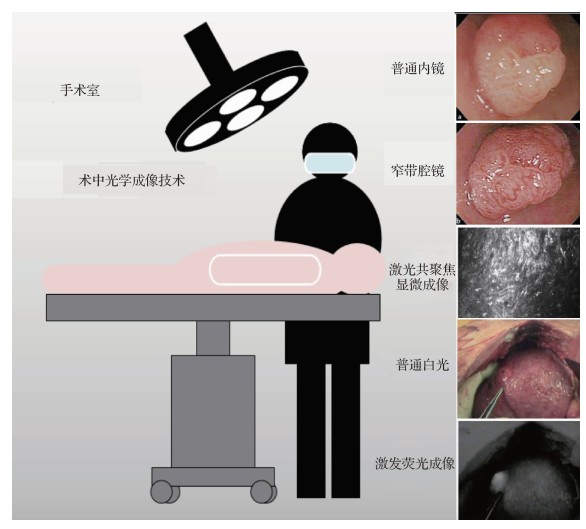 摘要:The rise of intraoperative optical imaging technologies provides a convenient and intuitive observation method for clinical surgery. Traditional intraoperative optical imaging methods include open optical and intraoperative endoscopic imaging. These methods ensure the smooth implementation of clinical surgery and promote the development of minimally invasive surgery. Subsequent methods include narrow-band endoscopic, intraoperative laser confocal microscopy, and near-infrared excited fluorescence imaging. Narrow-band endoscopic imaging uses a filter to filter out the broad-band spectrum emitted by the endoscope light source, leaving only the narrow-band spectrum for the diagnosis of various diseases of the digestive tract. The narrow-band spectrum is conducive to enhancing the image of the gastrointestinal mucosa vessels. In some lesions with microvascular changes, the narrow-band imaging system has evident advantages over ordinary endoscopy in distinguishing lesions. Narrow-band endoscopic imaging has also been widely used in the fields of otolaryngology, respiratory tract, gynecological endoscopy, and laparoscopic surgery in addition to the digestive tract. Intraoperative laser confocal microscopy is a new type of imaging method. It can realize superficial tissue imaging in vivo and provide pathological information by using the principle of excited fluorescence imaging. This imaging method has high clarity due to the application of confocal imaging and can be used for lesion positioning. Near-infrared excited fluorescence imaging uses excitation fluorescence imaging equipment combined with corresponding fluorescent contrast agents (such as ICG(indocyanine green), and methylene blue) to achieve intraoperative specific imaging of lesions, tissues, and organs in vivo. The basic principle is to stimulate the contrast agent accumulated in the tissue, the fluorescent contrast agent emits a fluorescent signal, and real-time imaging is realized by collecting the signals. In clinical research, the near-infrared fluorescence imaging technology is often used for lymphatic vessel tracing and accurate tumor resection. Contrast agents have different imaging spectral bands; hence, the corresponding near-infrared fluorescence imaging equipment is also developing to a multichannel imaging mode to image substantial contrast agents and label multiple tissues in the same field of view specifically during surgery. Multichannel near-infrared fluorescent surgical navigation equipment that has been gradually developed can realize simultaneous fluorescence imaging of multiple organs and tissues. These intraoperative optical imaging technologies can assist doctors in accurately locating tumors, rapidly distinguishing between benign and malignant tissues, and detecting small lesions. They have gained benefits in many clinical applications. However, optical imaging is susceptible to interference from ambient light, and optical signals are difficult to propagate in tissues without optical signal absorption and scattering. Intraoperative optical imaging technologies have the problems of limited imaging quality and superficial tissue imaging. In clinical research, intelligent analysis of preoperative imaging is fiercely developing, while information analysis of intraoperative imaging is still lacking of powerful analytical tools and analytical methods. The study of effective intraoperative optical imaging analysis algorithms needs further exploration. Machine learning is a tool developed with the age of computer information technology and is expected to provide an effective solution to the abovementioned problems. With the accumulation and explosion of data volume, deep learning, as a type of machine learning, is an end-to-end algorithm. It can gain the internal relationship among things autonomously through network training, establish an empirical model, and realize the function of traditional algorithms. Deep learning has shown enhanced results in the analysis and processing of natural images and is being continuously promoted and applied to various fields. Machine learning provides powerful technical means for intelligent analysis, image processing, and three-dimensional reconstruction, but the application research of using machine learning in intraoperative optical imaging is relatively few. The addition of machine learning is expected to break through the bottleneck and promote the development of intraoperative optical imaging technologies. This article focuses on intraoperative optical imaging technologies and investigates the application of machine learning in this field in recent years, including optimizing intraoperative optical imaging quality, assisting intelligent analysis of intraoperative optical imaging, and promoting three-dimensional modeling of intraoperative optical imaging. In the field of machine learning for intraoperative optical imaging optimization, existing research includes target detection of specific tissues, such as soft tissue segmentation and image fusion, and optimization of imaging effects, such as resolution enhancement of near-infrared fluorescence imaging during surgery and intraoperative endoscopic smoke removal. Furthermore, machine learning assists doctors in performing intraoperative optical imaging analysis, including the identification of benign and malignant tissues and the classification of lesion types and grades. Therefore, it can provide a timely reference value for the surgeon to judge the state of the patient during the clinical operation and before the pathological examination. In the field of intraoperative optical imaging reconstruction, machine learning can be combined with preoperative images (such as computed tomography and magnetic resonance imaging) to assist in intraoperative soft tissue reconstruction, or it can be based on intraoperative images for three-dimensional reconstruction. It can be used for localization, three-dimensional organ morphology reconstruction, and tracking of intraoperative tissues and surgical instruments. Thus, machine learning is expected to provide corresponding technical foundation for robotic surgery and augmented reality surgery in the future.This article summarizes and analyzes the application of machine learning in the field of intraoperative optical imaging and describes the application prospects of deep learning. As a review, it investigates the application research of machine learning in intraoperative optical imaging mainly from three aspects: intraoperative optical image optimization, intelligent analysis of optical imaging, and three-dimensional reconstruction. We also introduce related research and expected effects in the above fields. At the end of this article, the application of machine learning in the field of intraoperative optical imaging technologies is discussed, and the advantages and possible problems of machine-learning methods are analyzed. Furthermore, this article elaborates the possible future development direction of intraoperative optical imaging combined with machine learning, providing a broad view for subsequent research.关键词:intraoperative optical imaging;machine learning;imaging optimization;intelligent imaging analysis;3D modeling29|22|0更新时间:2024-05-07
摘要:The rise of intraoperative optical imaging technologies provides a convenient and intuitive observation method for clinical surgery. Traditional intraoperative optical imaging methods include open optical and intraoperative endoscopic imaging. These methods ensure the smooth implementation of clinical surgery and promote the development of minimally invasive surgery. Subsequent methods include narrow-band endoscopic, intraoperative laser confocal microscopy, and near-infrared excited fluorescence imaging. Narrow-band endoscopic imaging uses a filter to filter out the broad-band spectrum emitted by the endoscope light source, leaving only the narrow-band spectrum for the diagnosis of various diseases of the digestive tract. The narrow-band spectrum is conducive to enhancing the image of the gastrointestinal mucosa vessels. In some lesions with microvascular changes, the narrow-band imaging system has evident advantages over ordinary endoscopy in distinguishing lesions. Narrow-band endoscopic imaging has also been widely used in the fields of otolaryngology, respiratory tract, gynecological endoscopy, and laparoscopic surgery in addition to the digestive tract. Intraoperative laser confocal microscopy is a new type of imaging method. It can realize superficial tissue imaging in vivo and provide pathological information by using the principle of excited fluorescence imaging. This imaging method has high clarity due to the application of confocal imaging and can be used for lesion positioning. Near-infrared excited fluorescence imaging uses excitation fluorescence imaging equipment combined with corresponding fluorescent contrast agents (such as ICG(indocyanine green), and methylene blue) to achieve intraoperative specific imaging of lesions, tissues, and organs in vivo. The basic principle is to stimulate the contrast agent accumulated in the tissue, the fluorescent contrast agent emits a fluorescent signal, and real-time imaging is realized by collecting the signals. In clinical research, the near-infrared fluorescence imaging technology is often used for lymphatic vessel tracing and accurate tumor resection. Contrast agents have different imaging spectral bands; hence, the corresponding near-infrared fluorescence imaging equipment is also developing to a multichannel imaging mode to image substantial contrast agents and label multiple tissues in the same field of view specifically during surgery. Multichannel near-infrared fluorescent surgical navigation equipment that has been gradually developed can realize simultaneous fluorescence imaging of multiple organs and tissues. These intraoperative optical imaging technologies can assist doctors in accurately locating tumors, rapidly distinguishing between benign and malignant tissues, and detecting small lesions. They have gained benefits in many clinical applications. However, optical imaging is susceptible to interference from ambient light, and optical signals are difficult to propagate in tissues without optical signal absorption and scattering. Intraoperative optical imaging technologies have the problems of limited imaging quality and superficial tissue imaging. In clinical research, intelligent analysis of preoperative imaging is fiercely developing, while information analysis of intraoperative imaging is still lacking of powerful analytical tools and analytical methods. The study of effective intraoperative optical imaging analysis algorithms needs further exploration. Machine learning is a tool developed with the age of computer information technology and is expected to provide an effective solution to the abovementioned problems. With the accumulation and explosion of data volume, deep learning, as a type of machine learning, is an end-to-end algorithm. It can gain the internal relationship among things autonomously through network training, establish an empirical model, and realize the function of traditional algorithms. Deep learning has shown enhanced results in the analysis and processing of natural images and is being continuously promoted and applied to various fields. Machine learning provides powerful technical means for intelligent analysis, image processing, and three-dimensional reconstruction, but the application research of using machine learning in intraoperative optical imaging is relatively few. The addition of machine learning is expected to break through the bottleneck and promote the development of intraoperative optical imaging technologies. This article focuses on intraoperative optical imaging technologies and investigates the application of machine learning in this field in recent years, including optimizing intraoperative optical imaging quality, assisting intelligent analysis of intraoperative optical imaging, and promoting three-dimensional modeling of intraoperative optical imaging. In the field of machine learning for intraoperative optical imaging optimization, existing research includes target detection of specific tissues, such as soft tissue segmentation and image fusion, and optimization of imaging effects, such as resolution enhancement of near-infrared fluorescence imaging during surgery and intraoperative endoscopic smoke removal. Furthermore, machine learning assists doctors in performing intraoperative optical imaging analysis, including the identification of benign and malignant tissues and the classification of lesion types and grades. Therefore, it can provide a timely reference value for the surgeon to judge the state of the patient during the clinical operation and before the pathological examination. In the field of intraoperative optical imaging reconstruction, machine learning can be combined with preoperative images (such as computed tomography and magnetic resonance imaging) to assist in intraoperative soft tissue reconstruction, or it can be based on intraoperative images for three-dimensional reconstruction. It can be used for localization, three-dimensional organ morphology reconstruction, and tracking of intraoperative tissues and surgical instruments. Thus, machine learning is expected to provide corresponding technical foundation for robotic surgery and augmented reality surgery in the future.This article summarizes and analyzes the application of machine learning in the field of intraoperative optical imaging and describes the application prospects of deep learning. As a review, it investigates the application research of machine learning in intraoperative optical imaging mainly from three aspects: intraoperative optical image optimization, intelligent analysis of optical imaging, and three-dimensional reconstruction. We also introduce related research and expected effects in the above fields. At the end of this article, the application of machine learning in the field of intraoperative optical imaging technologies is discussed, and the advantages and possible problems of machine-learning methods are analyzed. Furthermore, this article elaborates the possible future development direction of intraoperative optical imaging combined with machine learning, providing a broad view for subsequent research.关键词:intraoperative optical imaging;machine learning;imaging optimization;intelligent imaging analysis;3D modeling29|22|0更新时间:2024-05-07
Review
-
 摘要:Medical imaging is an important tool used for medical diagnosis and clinical decision support that enables clinicians to view the internal of human bodies. Medical image analysis, as an important part of healthcare artificial intelligence, provides fast, smart, and accurate decision supports for clinicians and radiologists. 3D computer vision is an emerging research area with the rapid development and popularization of 3D sensors (e.g., light detection and ranging (LIDAR), RGB-D cameras) and computer-aided design in game industry and smart manufacturing. In particular, we focus on the interface of medical image analysis and 3D computer vision called medical 3D computer vision. We introduce the research advances and challenges in medical 3D computer vision in three levels, namely, tasks (medical 3D computer vision tasks), data (data modalities and datasets), and representation (efficient and effective representation learning for 3D images). First, we introduce classification, segmentation, detection, registration, and reconstruction in medical 3D computer vision at the task level. Classification, such as malignancy stratification and symptom estimation, is an everyday task for clinicians and radiologists. Segmentation denotes assigning each voxel (pixel) a semantic label. Detection refers to localizing key objects from medical images. Segmentation and detection include organ segmentation/detection and lesion segmentation/detection. Registration, that is, calculating the spatial transformation from one image to another, plays an important role in medical imaging scenarios, such as spatially aligning multiple images from serial examination of a follow-up patient. Reconstruction is also a key task in medical imaging that aims at fast and accurate imaging results to reduce patients' costs. Second, we introduce the important data modalities in medical 3D computer vision, such as computed tomography (CT), magnetic resonance imaging (MRI), and positron emission tomography (PET), at the data level. The principle and clinical scenario of each imaging modality are briefly discussed. We then depict a comprehensive list of medical 3D image research datasets that cover classification, segmentation, detection, registration, and reconstruction tasks in CT, MRI, and graphics format (mesh). Third, we discuss the representation learning for medical 3D computer vision. 2D convolutional neural networks, 3D convolutional neural networks, and hybrid approaches are the commonly used methods for 3D representation learning. 2D approaches can benefiting from large-scale 2D pretraining, triplanar, and trislice 2D representation for 3D medical images, whereas they are generally weak in capturing large 3D contexts. 3D approaches are natively strong in 3D context. However, few publicly available 3D medical datasets are large and sufficiently diverse for universal 3D pretraining. For hybrid (2D + 3D) approaches, we introduce multistream and multistage approaches. Although they are empirically effective, the intrinsic disadvantages within the 2D/3D parts still exist. To address the small-data issues for medical 3D computer vision, we discuss the pretraining approaches for medical 3D images. Pretraining for 3D convolutional neural network(CNN) with videos is straightforward to implement. However, a significant domain gap is found between medical images and videos. Collecting massive medical datasets for pretraining is theoretically feasible. However, it only results in thousands of 3D medical image cases with tens of medical datasets, which is significantly smaller compared with natural 2D image datasets. Research efforts exploring unsupervised (self-supervised) learning to obtain the pretrained 3D models are reported. Although its results are extremely impressive, the model performance of up-to-date unsupervised learning is incomparable with that of fully supervised learning. The unsupervised representation learning from medical 3D images cannot leverage the power of massive 2D supervised learning datasets. We introduce several techniques for 2D-to-3D transfer learning, including inflated 3D(I3D), axial-coronal-sagittal(ACS) convolutions, and AlignShift. I3D enables 2D-to-3D transfer learning by inflating 2D convolution kernels into 3D, and ACS convolutions and AlignShift enable that by introducing novel operators that shuffle the features from 3D receptive fields into a 2D manner. Finally, we discuss several research challenges, problems, and directions for medical 3D computer vision. We first determine the anisotropy issue in medical 3D images, which can be a source of domain gap, that is, between thick- and thin-slice data. We then discuss the data privacy and information silos in medical images, which are important factors that lead to small-data issues in medical 3D computer vision. Federated learning is highlighted as a possible solution for information silos. However, numerous problems, such as how to develop efficient systems and algorithms for federated learning, how to deal with adversarial participators in federated learning, and how to deal with unaligned and missing data, are found. We determine the data imbalance and long tail issues in medical 3D computer vision. Efficient and effective learning of representation from the noisy, imbalanced, and long-tailed real-world data can be extremely challenging in practice because of the imbalanced and long-tailed distributions of real-world patients. We mention the automatic machine learning as a future direction of medical 3D computer vision. With end-to-end deep learning, the development and deployment of medical image application is inapplicable. However, excessive engineering staff need to be tuned for a new medical image task, such as design of deep neural networks, choices of data argumentation, how to preform data preprocessing, and how to tune the learning procedure. The tuning of these hyperparameters can be performed with a hand-crafted or intelligent system to reduce the research efforts by numerous researchers and engineers. Thus, medical 3D computer vision is an emerging research area. With increasing large-scale datasets, easy-to-use and reproducible methodology, and innovative tasks, medical 3D computer vision is an exciting research area that can facilitate healthcare into a novel level.关键词:medical image analysis;3D computer vision;deep learning;convolutional neural networks(CNN);pre-training158|180|2更新时间:2024-05-07
摘要:Medical imaging is an important tool used for medical diagnosis and clinical decision support that enables clinicians to view the internal of human bodies. Medical image analysis, as an important part of healthcare artificial intelligence, provides fast, smart, and accurate decision supports for clinicians and radiologists. 3D computer vision is an emerging research area with the rapid development and popularization of 3D sensors (e.g., light detection and ranging (LIDAR), RGB-D cameras) and computer-aided design in game industry and smart manufacturing. In particular, we focus on the interface of medical image analysis and 3D computer vision called medical 3D computer vision. We introduce the research advances and challenges in medical 3D computer vision in three levels, namely, tasks (medical 3D computer vision tasks), data (data modalities and datasets), and representation (efficient and effective representation learning for 3D images). First, we introduce classification, segmentation, detection, registration, and reconstruction in medical 3D computer vision at the task level. Classification, such as malignancy stratification and symptom estimation, is an everyday task for clinicians and radiologists. Segmentation denotes assigning each voxel (pixel) a semantic label. Detection refers to localizing key objects from medical images. Segmentation and detection include organ segmentation/detection and lesion segmentation/detection. Registration, that is, calculating the spatial transformation from one image to another, plays an important role in medical imaging scenarios, such as spatially aligning multiple images from serial examination of a follow-up patient. Reconstruction is also a key task in medical imaging that aims at fast and accurate imaging results to reduce patients' costs. Second, we introduce the important data modalities in medical 3D computer vision, such as computed tomography (CT), magnetic resonance imaging (MRI), and positron emission tomography (PET), at the data level. The principle and clinical scenario of each imaging modality are briefly discussed. We then depict a comprehensive list of medical 3D image research datasets that cover classification, segmentation, detection, registration, and reconstruction tasks in CT, MRI, and graphics format (mesh). Third, we discuss the representation learning for medical 3D computer vision. 2D convolutional neural networks, 3D convolutional neural networks, and hybrid approaches are the commonly used methods for 3D representation learning. 2D approaches can benefiting from large-scale 2D pretraining, triplanar, and trislice 2D representation for 3D medical images, whereas they are generally weak in capturing large 3D contexts. 3D approaches are natively strong in 3D context. However, few publicly available 3D medical datasets are large and sufficiently diverse for universal 3D pretraining. For hybrid (2D + 3D) approaches, we introduce multistream and multistage approaches. Although they are empirically effective, the intrinsic disadvantages within the 2D/3D parts still exist. To address the small-data issues for medical 3D computer vision, we discuss the pretraining approaches for medical 3D images. Pretraining for 3D convolutional neural network(CNN) with videos is straightforward to implement. However, a significant domain gap is found between medical images and videos. Collecting massive medical datasets for pretraining is theoretically feasible. However, it only results in thousands of 3D medical image cases with tens of medical datasets, which is significantly smaller compared with natural 2D image datasets. Research efforts exploring unsupervised (self-supervised) learning to obtain the pretrained 3D models are reported. Although its results are extremely impressive, the model performance of up-to-date unsupervised learning is incomparable with that of fully supervised learning. The unsupervised representation learning from medical 3D images cannot leverage the power of massive 2D supervised learning datasets. We introduce several techniques for 2D-to-3D transfer learning, including inflated 3D(I3D), axial-coronal-sagittal(ACS) convolutions, and AlignShift. I3D enables 2D-to-3D transfer learning by inflating 2D convolution kernels into 3D, and ACS convolutions and AlignShift enable that by introducing novel operators that shuffle the features from 3D receptive fields into a 2D manner. Finally, we discuss several research challenges, problems, and directions for medical 3D computer vision. We first determine the anisotropy issue in medical 3D images, which can be a source of domain gap, that is, between thick- and thin-slice data. We then discuss the data privacy and information silos in medical images, which are important factors that lead to small-data issues in medical 3D computer vision. Federated learning is highlighted as a possible solution for information silos. However, numerous problems, such as how to develop efficient systems and algorithms for federated learning, how to deal with adversarial participators in federated learning, and how to deal with unaligned and missing data, are found. We determine the data imbalance and long tail issues in medical 3D computer vision. Efficient and effective learning of representation from the noisy, imbalanced, and long-tailed real-world data can be extremely challenging in practice because of the imbalanced and long-tailed distributions of real-world patients. We mention the automatic machine learning as a future direction of medical 3D computer vision. With end-to-end deep learning, the development and deployment of medical image application is inapplicable. However, excessive engineering staff need to be tuned for a new medical image task, such as design of deep neural networks, choices of data argumentation, how to preform data preprocessing, and how to tune the learning procedure. The tuning of these hyperparameters can be performed with a hand-crafted or intelligent system to reduce the research efforts by numerous researchers and engineers. Thus, medical 3D computer vision is an emerging research area. With increasing large-scale datasets, easy-to-use and reproducible methodology, and innovative tasks, medical 3D computer vision is an exciting research area that can facilitate healthcare into a novel level.关键词:medical image analysis;3D computer vision;deep learning;convolutional neural networks(CNN);pre-training158|180|2更新时间:2024-05-07 -
 摘要:A point cloud refers to a set of data points in a three-dimensional space. Each point is composed of a three-dimensional coordinate, with object's reflectivity, reflection intensity, distance from the point to the center of the scanner, horizontal angle, vertical angle, and deviation value. The point cloud is obtained by two methods, one is obtained by scanning the target object by the three-dimensional sensing device, such as LiDAR sensor and RGB-D camera, and the other is obtained by reconstruction from two-dimensional medical images. The point cloud can express the geometric position, shape, and scale of the target object. The point cloud has a wide range of applications in areas such as autonomous driving, robots, surveillance systems, surveying and mapping geography, virtual reality, and medicine, which has achieved remarkable results. Many researchers in the field of medical imaging have also devoted themselves to the research of medical image point cloud processing algorithms. Point cloud can intuitively simulate the three-dimensional structure of biological organs and tissues. With important application value in clinical medicine, the classification, segmentation, registration, and other tasks based on medical point cloud can help doctors make accurate diagnosis and treatment. The point cloud-based medical diagnosis has advantages and has the potential for future application in clinical screening diagnosis, personalized medical device-assisted design, and 3D printing. At this stage, deep learning algorithms have achieved remarkable results in tasks such as target detection, segmentation, and recognition. Deep learning algorithms gradually become efficient and popular in tasks such as target detection, segmentation, and recognition. Therefore, increasing point cloud processing algorithms are gradually extended from traditional algorithms to deep learning algorithms. This article reviews the research and progress of point cloud algorithms in the medical field. This review aims to summarize the current point cloud methods used in the medical field and focuses on 1) the characteristics, acquisition methods, and data conversion methods of medical point clouds; 2) traditional algorithms and deep learning algorithms in medical point cloud segmentation; and 3) the definition and significance of medical point cloud registration tasks. This review is based on feature or non-feature registration method. Finally, although the point cloud method has been applied in the medical field, the current application of the point cloud-based frontier method in the medical point cloud is still insufficient. Applying state-of-the-art algorithms to medical point clouds also requires continuous in-depth exploration and research. To date, medical point clouds have been used to assist doctors in completing some diagnostic tasks, but they are still in the process of continuous development and cannot replace the role of clinicians. The clinical application of medical point cloud has some limitations and challenges: 1) In the application research of point clouds in the medical field, the first task is to obtain point cloud data that can accurately characterize disease information. At present, point cloud data acquisition methods are relatively simple. In the future, high-quality point cloud imaging equipment can be combined to obtain accurate medical point cloud dataset. In applied research, the first task is to obtain point cloud data that can accurately characterize medical anatomical structure information. Considering that the morphological structure of human tissues and organs is relatively complex, most of the point cloud data of human organs are obtained by reconstructing medical images (such as (computed tomography(CT)) and (magnetic resonance imaging(MRI)). Therefore, such point clouds are sparsely distributed, with noise and errors. Obtaining accurate and dense medical point cloud datasets from medical images is an important subject to be studied. 2) In addition to facing the challenges of sparse reconstruction and data imbalance in point clouds, the difficulty of labeling medical point cloud data sets, the high cost of data integration, and the inevitable subjective labeling errors are the reasons why deep learning algorithms have not been widely used in the field of medical point clouds. The small amount of sample data and the imbalance of sample data may affect the accuracy of disease diagnosis. In the future, methods such as semi-supervised learning, active learning, and generating samples against the generated network can be used to improve learning accuracy. 3) A large number of medical point clouds are generated in the hospital but are not used to train and improve the diagnostic model. With the emergence of super-resolution algorithms and point cloud up-sampling networks, the prediction of sparse point clouds to dense point clouds based on medical image reconstruction will be an important means to construct high-quality medical point clouds. In the future, with the improvement of the quality and quantity of medical point cloud data sets, the research of medical point cloud processing algorithms will attract more researchers. Current research only focuses on model training and evaluation of specific data sets, which makes the universality of these algorithms challenging. The application and development of point cloud in medical images are currently a hot topic. Although point clouds have gradually penetrated into a considerable number of fields in medicine, the application of the current frontier methods of point cloud processing in medical point clouds is still insufficient. Research work using medical point cloud still needs to invest more research energy and attention.关键词:point clouds;medical applications;deep learning;segmentation;registration213|80|8更新时间:2024-05-07
摘要:A point cloud refers to a set of data points in a three-dimensional space. Each point is composed of a three-dimensional coordinate, with object's reflectivity, reflection intensity, distance from the point to the center of the scanner, horizontal angle, vertical angle, and deviation value. The point cloud is obtained by two methods, one is obtained by scanning the target object by the three-dimensional sensing device, such as LiDAR sensor and RGB-D camera, and the other is obtained by reconstruction from two-dimensional medical images. The point cloud can express the geometric position, shape, and scale of the target object. The point cloud has a wide range of applications in areas such as autonomous driving, robots, surveillance systems, surveying and mapping geography, virtual reality, and medicine, which has achieved remarkable results. Many researchers in the field of medical imaging have also devoted themselves to the research of medical image point cloud processing algorithms. Point cloud can intuitively simulate the three-dimensional structure of biological organs and tissues. With important application value in clinical medicine, the classification, segmentation, registration, and other tasks based on medical point cloud can help doctors make accurate diagnosis and treatment. The point cloud-based medical diagnosis has advantages and has the potential for future application in clinical screening diagnosis, personalized medical device-assisted design, and 3D printing. At this stage, deep learning algorithms have achieved remarkable results in tasks such as target detection, segmentation, and recognition. Deep learning algorithms gradually become efficient and popular in tasks such as target detection, segmentation, and recognition. Therefore, increasing point cloud processing algorithms are gradually extended from traditional algorithms to deep learning algorithms. This article reviews the research and progress of point cloud algorithms in the medical field. This review aims to summarize the current point cloud methods used in the medical field and focuses on 1) the characteristics, acquisition methods, and data conversion methods of medical point clouds; 2) traditional algorithms and deep learning algorithms in medical point cloud segmentation; and 3) the definition and significance of medical point cloud registration tasks. This review is based on feature or non-feature registration method. Finally, although the point cloud method has been applied in the medical field, the current application of the point cloud-based frontier method in the medical point cloud is still insufficient. Applying state-of-the-art algorithms to medical point clouds also requires continuous in-depth exploration and research. To date, medical point clouds have been used to assist doctors in completing some diagnostic tasks, but they are still in the process of continuous development and cannot replace the role of clinicians. The clinical application of medical point cloud has some limitations and challenges: 1) In the application research of point clouds in the medical field, the first task is to obtain point cloud data that can accurately characterize disease information. At present, point cloud data acquisition methods are relatively simple. In the future, high-quality point cloud imaging equipment can be combined to obtain accurate medical point cloud dataset. In applied research, the first task is to obtain point cloud data that can accurately characterize medical anatomical structure information. Considering that the morphological structure of human tissues and organs is relatively complex, most of the point cloud data of human organs are obtained by reconstructing medical images (such as (computed tomography(CT)) and (magnetic resonance imaging(MRI)). Therefore, such point clouds are sparsely distributed, with noise and errors. Obtaining accurate and dense medical point cloud datasets from medical images is an important subject to be studied. 2) In addition to facing the challenges of sparse reconstruction and data imbalance in point clouds, the difficulty of labeling medical point cloud data sets, the high cost of data integration, and the inevitable subjective labeling errors are the reasons why deep learning algorithms have not been widely used in the field of medical point clouds. The small amount of sample data and the imbalance of sample data may affect the accuracy of disease diagnosis. In the future, methods such as semi-supervised learning, active learning, and generating samples against the generated network can be used to improve learning accuracy. 3) A large number of medical point clouds are generated in the hospital but are not used to train and improve the diagnostic model. With the emergence of super-resolution algorithms and point cloud up-sampling networks, the prediction of sparse point clouds to dense point clouds based on medical image reconstruction will be an important means to construct high-quality medical point clouds. In the future, with the improvement of the quality and quantity of medical point cloud data sets, the research of medical point cloud processing algorithms will attract more researchers. Current research only focuses on model training and evaluation of specific data sets, which makes the universality of these algorithms challenging. The application and development of point cloud in medical images are currently a hot topic. Although point clouds have gradually penetrated into a considerable number of fields in medicine, the application of the current frontier methods of point cloud processing in medical point clouds is still insufficient. Research work using medical point cloud still needs to invest more research energy and attention.关键词:point clouds;medical applications;deep learning;segmentation;registration213|80|8更新时间:2024-05-07 -
 摘要:Hepatocellular carcinoma is one of the most common malignant tumors of the digestive system in clinic. It ranks third after gastric cancer and lung cancer in the death ranking of malignant tumors. Computed tomography (CT) can well display the organs composed of soft tissue and show the lesions in the abdominal image. It has become a typical method for the diagnosis and treatment of liver diseases. It produces high-quality liver imaging that can provide comprehensive information for the diagnosis and treatment of liver tumors, alleviate the heavy workload of doctors, and have an important value for subsequent diagnosis and treatment. Segmentation of CT images of liver tumors is a crucial step in the diagnosis of liver cancer. In accordance with the maximum diameter, volume, and number of liver lesions, medical workers can give patients accurate diagnosis results and treatment plans conveniently and rapidly. However, the manual three-dimensional segmentation of liver tumors is time consuming and requires substantial work. Therefore, a method for automatically segmenting liver tumors is urgently needed. Many challenges occur in the segmentation of liver tumors. First, the CT image of a liver tumor shows the cross section of the human body, and the contrast of the liver and liver tumor tissue is inconsiderably different from that of the surrounding adjacent tissues (such as the stomach, pancreas, and heart). The segmentation by using grayscale differences is difficult. Second, the individual differences of patients result in diverse sizes and shapes of liver tumors. Third, CT images are susceptible to various external factors, such as noise, partial volume effects, and magnetic field bias. The interference of the shift makes the image blurry. Dealing with the effects of these factors in a timely manner is a great challenge for medical imaging researchers. Accurate segmentation can ensure that clinicians can make wise surgical treatment plans. With the rise of big data and artificial intelligence in recent years, assisted diagnosis of liver cancer based on deep learning has gradually become a popular research topic. Its combination with medicine can realize and predict the condition and assist diagnosis, which has great clinical significance. Segmentation methods for liver tumor CT images based on deep learning have also attracted wide attention in the past few years. From relevant literature in the field of liver tumor image segmentation, this paper mainly summarizes several commonly used segmentation methods for current liver tumor CT images based on deep learning, aiming to provide convenience to related researchers. We comprehensively summarize and analyze the deep learning methods for liver tumor CT images from three aspects: datasets, evaluation indicators and algorithms. First, we introduce common databases of liver tumors and analyze and compare them in terms of year, resolution, number of cases, slice thickness, pixel size, and voxel size to compare the segmentation methods for emerging liver tumors objectively. Second, several important evaluation indicators, such as Dice, relative volume difference, and volumetric overlap error, are also briefly introduced, analyzed, and compared to evaluate the effectiveness of each algorithm in the accuracy of liver tumor segmentation. On the basis of the previous work, we divide the deep learning segmentation methods for CT images of liver tumors into three categories, namely, liver tumor segmentation methods based on fully convolutional network (FCN), U-Net, and generative adversarial network (GAN). The segmentation methods based on FCN can be further divided into two- and three-dimensional methods in accordance with the dimension of the convolution kernel. The segmentation methods based on U-Net are divided into three subcategories, which are methods based on single network, methods based on multinetwork, and methods combined with traditional methods. Similarly, the segmentation methods based on GAN are divided into three subcategories, which are based on network architecture improvements, generator-based improvements, and other methods. The basic ideas, network architecture forms, improvement schemes, advantages, and disadvantages of various methods are emphasized, and the performance of these methods on typical datasets is compared. Lastly, the advantages, disadvantages, and application scope of the three methods are summarized and compared. The future research trends of liver tumor deep learning segmentation methods are analyzed. 1) The use of three-dimensional neural networks and network deepening is a future research direction in this field. 2) The use of multimodal liver images for segmentation and the combination of multiple different deep neural networks to extract deep information of images for improving the accuracy of liver tumor segmentation are also main research directions in this field. 3) To overcome the problem of lack or unavailability of data, some researchers have shifted the supervised field to a semi-supervised or unsupervised field. For example, GAN is combined with other higher-performance networks. This situation can be further studied in the future. In summary, accurate segmentation of liver tumors is a necessary step in liver disease diagnosis, surgical planning, and postoperative evaluation. Deep learning is superior to traditional segmentation methods when segmenting liver tumors, and the obtained images have higher sensitivity and specificity. This study hopes that clinicians can intuitively and clearly observe the anatomical structure of normal and diseased tissues through the increasingly mature liver tumor segmentation technologies. It provides a scientific basis for clinical diagnosis, surgical procedures, and biomedical research. The research and development of medical image segmentation technologies play an important role in the reform of the medical field and have great research value and significance.关键词:computed tomography(CT);liver tumor;deep learning;medical image segmentation;fully convolutional network(FCN);U-Net;generative adversarial network(GAN)235|78|9更新时间:2024-05-07
摘要:Hepatocellular carcinoma is one of the most common malignant tumors of the digestive system in clinic. It ranks third after gastric cancer and lung cancer in the death ranking of malignant tumors. Computed tomography (CT) can well display the organs composed of soft tissue and show the lesions in the abdominal image. It has become a typical method for the diagnosis and treatment of liver diseases. It produces high-quality liver imaging that can provide comprehensive information for the diagnosis and treatment of liver tumors, alleviate the heavy workload of doctors, and have an important value for subsequent diagnosis and treatment. Segmentation of CT images of liver tumors is a crucial step in the diagnosis of liver cancer. In accordance with the maximum diameter, volume, and number of liver lesions, medical workers can give patients accurate diagnosis results and treatment plans conveniently and rapidly. However, the manual three-dimensional segmentation of liver tumors is time consuming and requires substantial work. Therefore, a method for automatically segmenting liver tumors is urgently needed. Many challenges occur in the segmentation of liver tumors. First, the CT image of a liver tumor shows the cross section of the human body, and the contrast of the liver and liver tumor tissue is inconsiderably different from that of the surrounding adjacent tissues (such as the stomach, pancreas, and heart). The segmentation by using grayscale differences is difficult. Second, the individual differences of patients result in diverse sizes and shapes of liver tumors. Third, CT images are susceptible to various external factors, such as noise, partial volume effects, and magnetic field bias. The interference of the shift makes the image blurry. Dealing with the effects of these factors in a timely manner is a great challenge for medical imaging researchers. Accurate segmentation can ensure that clinicians can make wise surgical treatment plans. With the rise of big data and artificial intelligence in recent years, assisted diagnosis of liver cancer based on deep learning has gradually become a popular research topic. Its combination with medicine can realize and predict the condition and assist diagnosis, which has great clinical significance. Segmentation methods for liver tumor CT images based on deep learning have also attracted wide attention in the past few years. From relevant literature in the field of liver tumor image segmentation, this paper mainly summarizes several commonly used segmentation methods for current liver tumor CT images based on deep learning, aiming to provide convenience to related researchers. We comprehensively summarize and analyze the deep learning methods for liver tumor CT images from three aspects: datasets, evaluation indicators and algorithms. First, we introduce common databases of liver tumors and analyze and compare them in terms of year, resolution, number of cases, slice thickness, pixel size, and voxel size to compare the segmentation methods for emerging liver tumors objectively. Second, several important evaluation indicators, such as Dice, relative volume difference, and volumetric overlap error, are also briefly introduced, analyzed, and compared to evaluate the effectiveness of each algorithm in the accuracy of liver tumor segmentation. On the basis of the previous work, we divide the deep learning segmentation methods for CT images of liver tumors into three categories, namely, liver tumor segmentation methods based on fully convolutional network (FCN), U-Net, and generative adversarial network (GAN). The segmentation methods based on FCN can be further divided into two- and three-dimensional methods in accordance with the dimension of the convolution kernel. The segmentation methods based on U-Net are divided into three subcategories, which are methods based on single network, methods based on multinetwork, and methods combined with traditional methods. Similarly, the segmentation methods based on GAN are divided into three subcategories, which are based on network architecture improvements, generator-based improvements, and other methods. The basic ideas, network architecture forms, improvement schemes, advantages, and disadvantages of various methods are emphasized, and the performance of these methods on typical datasets is compared. Lastly, the advantages, disadvantages, and application scope of the three methods are summarized and compared. The future research trends of liver tumor deep learning segmentation methods are analyzed. 1) The use of three-dimensional neural networks and network deepening is a future research direction in this field. 2) The use of multimodal liver images for segmentation and the combination of multiple different deep neural networks to extract deep information of images for improving the accuracy of liver tumor segmentation are also main research directions in this field. 3) To overcome the problem of lack or unavailability of data, some researchers have shifted the supervised field to a semi-supervised or unsupervised field. For example, GAN is combined with other higher-performance networks. This situation can be further studied in the future. In summary, accurate segmentation of liver tumors is a necessary step in liver disease diagnosis, surgical planning, and postoperative evaluation. Deep learning is superior to traditional segmentation methods when segmenting liver tumors, and the obtained images have higher sensitivity and specificity. This study hopes that clinicians can intuitively and clearly observe the anatomical structure of normal and diseased tissues through the increasingly mature liver tumor segmentation technologies. It provides a scientific basis for clinical diagnosis, surgical procedures, and biomedical research. The research and development of medical image segmentation technologies play an important role in the reform of the medical field and have great research value and significance.关键词:computed tomography(CT);liver tumor;deep learning;medical image segmentation;fully convolutional network(FCN);U-Net;generative adversarial network(GAN)235|78|9更新时间:2024-05-07 - 摘要:Schizophrenia is a severe psychiatric disorder with abnormalities in brain structure and function. The early diagnosis and timely intervention can significantly improve the quality of life. However, no effective methods are available for the diagnosis and treatment of schizophrenia yet due to the complex pathology of this disease. Recently, deep learning methods have been widely used in medical imaging and have shown their great feature representation capability. The performance of deep learning methods has been demonstrated to be superior to that of traditional machine-learning methods. Researchers have attempted to use deep learning to assist the diagnosis of schizophrenia on the basis of magnetic resonance imaging (MRI) data. However, few surveys have systematically summarized the application of deep learning in the MRI-based diagnosis of schizophrenia. Therefore, MRI-based deep learning methods for the classification of schizophrenia patients and healthy controls were reviewed in this paper. First, commonly used deep learning models in the field of MRI-based diagnosis of schizophrenia were introduced. These models were classified into five categories: feed-forward neural network, recurrent neural network, convolutional neural network, unsupervised feature-learning models, and other deep models. The basic idea of the model in each category was introduced. Specifically, perceptron and its deeper expanding model, called multilayer perceptron, were briefly introduced in the review of feed-forward neural network. The most famous recurrent neural network model, named long short-term memory, was described. For the convolutional neural network, the common model components were reviewed. Unsupervised feature-learning models were further divided into two subcategories: stacked auto-encoder and deep belief network. The basic components of the model in each subcategory were described. We also reviewed two other deep models that cannot be classified into the categories described above, namely, capsule network and multi-grained cascade forest. Examples of deep learning studies used in the diagnosis of schizophrenia in each category were included to show the effectiveness of these models. Second, the deep learning methods for schizophrenia classification were divided on the basis of the data modality of MRI and reviewed in accordance with the following categories: structural MRI (sMRI)-, functional MRI (fMRI)-, and multimodal-based methods. Papers using MRI-based deep learning models for schizophrenia diagnosis were also summarized in a table. For sMRI-based methods, studies using 2D and 3D models and different methods for solving the requirement of large sample size were discussed. Resting-state fMRI- and task-fMRI-based methods were included in the fMRI-based category. In addition to different methods for solving limited sample size, studies using regional- or voxel-based functional activity features and functional connectivity-based features were introduced in this category. Some multimodal-based methods, which show better performance than most of unimodal-based approaches, were also described. Lastly, on the basis of existing studies, we concluded the main challenges and obstacles in the real application of these models in schizophrenia diagnosis. The first one refers to sample-related problems, including limited sample size and imbalanced sample class distribution. The second one is the lack of interpretability in deep learning models, which is a critical limitation for the application of deep learning models in real clinical condition. The last one is related to problems in multimodality analyses. Missing modality and the lack of effective multimodal feature fusion strategies would be the key problems to be solved. Future research directions lie in multisite and longitudinal data analyses and the construction of personalized precise models for the diagnosis of different subtypes of schizophrenia. Multisite data analyses would lead to a robust model and enable effective evaluation for deep learning models before the translation to clinical applications. Longitudinal data would provide the dynamically developmental patterns of the disease, leading to enhanced model performance. Personalized precise models are urgently needed to improve the accuracy of computer-aided diagnostic systems in consideration of the broad symptoms of schizophrenia and existing biased methods. In summary, this article reviewed the application of MRI-based deep learning models in the diagnosis of schizophrenia and provided guidance for future studies in this field.关键词:deep learning;schizophrenia;magnetic resonance imaging(MRI);diagnosis;classification68|92|0更新时间:2024-05-07
-
 摘要:In March 2020, the World Health Organization(WHO) declared the new corona virus pneumonia (COVID-19) as a world pandemic, which means that the epidemic has broken out worldwide. The outbreak of COVID-19 threatens the lives and property safety of countless people and brings great pressure to medical systems. The main clinical symptoms of COVID-19 are fever, cough, and fatigue, which may lead to a fatal complication: acute respiratory distress syndrome. The main challenge in inhibiting the spread of this disease is the lack of efficient detection methods. Although reverse transcription-polymerase chain reaction (RT-PCR) is the gold standard for confirming COVID-19, it takes 4-6 h to obtain the results, and the false-negative rate of RT-PCR detection is as high as 17%-25.5%. Therefore, multiple RT-PCR detections at intervals of several days must be performed to confirm the diagnostic result. In addition, RT-PCR reagents are lacking in many severe epidemic areas. By contrast, X-ray and CT(computed tomography) examination equipment have been widely popularized in hospitals. In clinical practice, by combining clinical symptoms and travel history, CT is an efficient and safe method to diagnose COVID-19. Compared with CT, X-ray examination has faster scanning speed and lower radiation amount. Moreover, X-ray and CT images are important tools for doctors to track and observe the condition and evaluate the efficacy. In summary, medical imaging plays a vital role in limiting the spread of viruses and treating COVID-19. During the outbreak of the epidemic, medical imaging-based AI-assisted diagnostic technology has become a popular research direction. Computer-aided diagnostic technology improves the sensitivity and specificity of doctors' diagnosis and is accurate and efficient, which helps rapid diagnosis of a large number of suspicious cases. For example, the out preformed AI-assisted diagnosis system can achieve an accuracy rate comparable to that of radiologists, and it take less than 1 second to perform a diagnosis. The system has been used in 16 hospitals, with more than 1 300 diagnoses performed daily. This article reviews the latest research works on AI-assisted diagnosis of COVID-19 and analyzes and summarizes them on the basis of four aspects: data preparation, image segmentation, diagnosis, and prognosis. First, this article organizes some public data sets to support the AI-assisted diagnostic technology of COVID-19 and provides several solutions to insufficient datasets, such as the human-in-the-loop strategy, which improves the efficiency of data set production. Using transfer learning, weakly supervised or unsupervised learning can reduce model's dependence on the COVID-19 dataset. Second, the semantic segmentation network is also an indispensable part of the intelligent diagnosis of COVID-19. Segmenting the lung region from the original image is a key pre-processing step, which can reduce the calculation amount of subsequent algorithms. The lesion area helps the doctor to track the condition of the disease, and the infection rate can be calculated according to the size of the infected area. U-Net, U-Net++, and attention U-Net are suitable for the segmentation of medical images because of the small number of parameters, which is not easy to overfit. Furthermore, training the semantic segmentation network with the idea of the generative adversarial network (GAN) can improve the Dice coefficient. Third, this article introduces the AI diagnostic system from two aspects of CT images and X-rays. Comparing different diagnostic schemes, the method of diagnosis based on the segmentation images is better than that based on the original images. Among the classification networks, ResNet and VGG19(visual geometry group 19-layer net) perform better. Methods such as GAN, location attention mechanisms, transfer learning, and combining 2D and 3D features can be used to improve accuracy. In addition, clinical information (travel and contact history, white blood cell count, fever, cough, sputum, patient age, and patient gender) can be used as a basis for diagnosis. For example, algorithm D_FF_Conic uses clinical information as a diagnostic basis and has reached an accuracy rate of 90%. Clinicians will consider medical imaging and clinical information in the process of diagnosis, but the current AI diagnostic system cannot integrate multiple types of data for diagnosis. Although some algorithms can fuse the diagnostic results of medical images with the diagnostic results of clinical information, the simple fine-tuned algorithm haven't learned the deep internal connection between different types of data. Fourth, AI technology can also predict high-risk patients on the basis of infection rates and clinical information. Some research predicted the survival rate of COVID-19 patients on the basis of age, syndrome, and infection rate. Such algorithms can help doctors find and treat high-risk patients early, thereby reducing mortality, which is of great significance. This article shows the latest progress of COVID-19's medical imaging-based AI diagnosis. Although some AI-assisted diagnostic systems have been deployed in hospitals to play a practical role, these algorithms still have some problems, such as insufficient training data, a single diagnostic basis, and the ability to distinguish between non-COVID-19 pneumonia and COVID-19.关键词:artificial intelligence;COVID-19;image segmentation;computer aided diagnosis;infection region segmentation191|214|3更新时间:2024-05-07
摘要:In March 2020, the World Health Organization(WHO) declared the new corona virus pneumonia (COVID-19) as a world pandemic, which means that the epidemic has broken out worldwide. The outbreak of COVID-19 threatens the lives and property safety of countless people and brings great pressure to medical systems. The main clinical symptoms of COVID-19 are fever, cough, and fatigue, which may lead to a fatal complication: acute respiratory distress syndrome. The main challenge in inhibiting the spread of this disease is the lack of efficient detection methods. Although reverse transcription-polymerase chain reaction (RT-PCR) is the gold standard for confirming COVID-19, it takes 4-6 h to obtain the results, and the false-negative rate of RT-PCR detection is as high as 17%-25.5%. Therefore, multiple RT-PCR detections at intervals of several days must be performed to confirm the diagnostic result. In addition, RT-PCR reagents are lacking in many severe epidemic areas. By contrast, X-ray and CT(computed tomography) examination equipment have been widely popularized in hospitals. In clinical practice, by combining clinical symptoms and travel history, CT is an efficient and safe method to diagnose COVID-19. Compared with CT, X-ray examination has faster scanning speed and lower radiation amount. Moreover, X-ray and CT images are important tools for doctors to track and observe the condition and evaluate the efficacy. In summary, medical imaging plays a vital role in limiting the spread of viruses and treating COVID-19. During the outbreak of the epidemic, medical imaging-based AI-assisted diagnostic technology has become a popular research direction. Computer-aided diagnostic technology improves the sensitivity and specificity of doctors' diagnosis and is accurate and efficient, which helps rapid diagnosis of a large number of suspicious cases. For example, the out preformed AI-assisted diagnosis system can achieve an accuracy rate comparable to that of radiologists, and it take less than 1 second to perform a diagnosis. The system has been used in 16 hospitals, with more than 1 300 diagnoses performed daily. This article reviews the latest research works on AI-assisted diagnosis of COVID-19 and analyzes and summarizes them on the basis of four aspects: data preparation, image segmentation, diagnosis, and prognosis. First, this article organizes some public data sets to support the AI-assisted diagnostic technology of COVID-19 and provides several solutions to insufficient datasets, such as the human-in-the-loop strategy, which improves the efficiency of data set production. Using transfer learning, weakly supervised or unsupervised learning can reduce model's dependence on the COVID-19 dataset. Second, the semantic segmentation network is also an indispensable part of the intelligent diagnosis of COVID-19. Segmenting the lung region from the original image is a key pre-processing step, which can reduce the calculation amount of subsequent algorithms. The lesion area helps the doctor to track the condition of the disease, and the infection rate can be calculated according to the size of the infected area. U-Net, U-Net++, and attention U-Net are suitable for the segmentation of medical images because of the small number of parameters, which is not easy to overfit. Furthermore, training the semantic segmentation network with the idea of the generative adversarial network (GAN) can improve the Dice coefficient. Third, this article introduces the AI diagnostic system from two aspects of CT images and X-rays. Comparing different diagnostic schemes, the method of diagnosis based on the segmentation images is better than that based on the original images. Among the classification networks, ResNet and VGG19(visual geometry group 19-layer net) perform better. Methods such as GAN, location attention mechanisms, transfer learning, and combining 2D and 3D features can be used to improve accuracy. In addition, clinical information (travel and contact history, white blood cell count, fever, cough, sputum, patient age, and patient gender) can be used as a basis for diagnosis. For example, algorithm D_FF_Conic uses clinical information as a diagnostic basis and has reached an accuracy rate of 90%. Clinicians will consider medical imaging and clinical information in the process of diagnosis, but the current AI diagnostic system cannot integrate multiple types of data for diagnosis. Although some algorithms can fuse the diagnostic results of medical images with the diagnostic results of clinical information, the simple fine-tuned algorithm haven't learned the deep internal connection between different types of data. Fourth, AI technology can also predict high-risk patients on the basis of infection rates and clinical information. Some research predicted the survival rate of COVID-19 patients on the basis of age, syndrome, and infection rate. Such algorithms can help doctors find and treat high-risk patients early, thereby reducing mortality, which is of great significance. This article shows the latest progress of COVID-19's medical imaging-based AI diagnosis. Although some AI-assisted diagnostic systems have been deployed in hospitals to play a practical role, these algorithms still have some problems, such as insufficient training data, a single diagnostic basis, and the ability to distinguish between non-COVID-19 pneumonia and COVID-19.关键词:artificial intelligence;COVID-19;image segmentation;computer aided diagnosis;infection region segmentation191|214|3更新时间:2024-05-07 -
 摘要:Emerging interest has been shown in the study of infant brain development by using magnetic resonance (MR) imaging because it provides a safe and noninvasive way of examining cross-sectional views of the brain in multiple contrasts. Quantitative analysis of brain MR images is a conventional routine for many neurological diseases and conditions, which relies on the accurate segmentation of structures of interest. Accurate segmentation of infant brain MR images into white matter (WM), gray matter (GM), and cerebrospinal fluid is of great importance in studying and measuring normal and abnormal early brain development. However, in the isointense phase (6-9 months of age), WM and GM exhibit similar levels of intensity in T1-weighted and T2-weighted MR images due to the inherent myelination and maturation, posing significant challenges for automated segmentation. Compared with traditional methods, deep learning-based methods have greatly improved the accuracy and efficiency of isointense infant brain segmentation. Thus, deep learning-based segmentation methods have been increasingly used by researchers due to their excellent performance. Nevertheless, no literature has systematically summarized and analyzed the methods in this field. The current study aims to review existing deep learning-based approaches for isointense infant brain MR image segmentation. With the extensive research in the literature, we systematically summarized the current deep learning-based methods for isointense infant brain MR image segmentation. We first introduced an authoritative isointense infant brain segmentation dataset, which was used in the iSeg-2017 challenge, hosted as a part of the Medical Image Computing and Computer Assisted Intervention Society Conference 2017. Afterward, several evaluation metrics, including Dice coefficient, 95th-percentile Hausdorff distance, and average symmetric surface distance, were briefly described. We classified the existing deep learning-based methods for isointense infant brain MR image segmentation into two categories: 2D and fully convolutional neural network-based methods. Fully convolutional neural network-based methods can be further divided into two subcategories: 2D and 3D network-based approaches. On the basis of the two categories, we comprehensively described and analyzed the basic ideas, network architecture, improvement schemes, and segmentation performance of each method. In addition, we compared the performance of some existing deep learning-based methods and summarized the analysis results of typical methods on the iSeg-2017 dataset. Lastly, three possible research directions of isointense infant brain MR image segmentation methods based on deep learning were discussed. We drew three main conclusions by reviewing the main work in this field, thereby providing a good overview of existing deep learning methods for isointense infant brain MR image segmentation. First, using multimodality data is beneficial for the network to obtain rich feature information, which can improve the segmentation accuracy. Second, compared with 2D fully convolutional network-based methods, 3D-based methods for MR image brain segmentation can integrate richer spatial context information, resulting in higher accuracy and efficiency. Third, adopting complicated network architecture and dense network connection could make the network achieve superior accuracy performance. Three possible future directions, namely, embedding powerful feature representation modules (e.g., attention mechanism), adding prior knowledge of the infant brain to the network model (e.g., the cortical thickness of infant brain is within a certain range), and constructing a semisupervised or weakly supervised network model trained with a small amount of labeled data, were recommended. Precise segmentation of infant brain tissues is an essential step towards comprehensive volumetric studies and quantitative analysis of early brain development. Deep learning has shown great advantages in isointense infant brain segmentation, and its accuracy and efficiency have been greatly improved compared with traditional methods. With the development of deep learning, it will bring further improvement in the research field of isointense infant brain segmentation and promote the research of early human brain development.关键词:magnetic resonance imaging;isointense phase;infant brain segmentation;deep learning;convolutional neural network101|135|4更新时间:2024-05-07
摘要:Emerging interest has been shown in the study of infant brain development by using magnetic resonance (MR) imaging because it provides a safe and noninvasive way of examining cross-sectional views of the brain in multiple contrasts. Quantitative analysis of brain MR images is a conventional routine for many neurological diseases and conditions, which relies on the accurate segmentation of structures of interest. Accurate segmentation of infant brain MR images into white matter (WM), gray matter (GM), and cerebrospinal fluid is of great importance in studying and measuring normal and abnormal early brain development. However, in the isointense phase (6-9 months of age), WM and GM exhibit similar levels of intensity in T1-weighted and T2-weighted MR images due to the inherent myelination and maturation, posing significant challenges for automated segmentation. Compared with traditional methods, deep learning-based methods have greatly improved the accuracy and efficiency of isointense infant brain segmentation. Thus, deep learning-based segmentation methods have been increasingly used by researchers due to their excellent performance. Nevertheless, no literature has systematically summarized and analyzed the methods in this field. The current study aims to review existing deep learning-based approaches for isointense infant brain MR image segmentation. With the extensive research in the literature, we systematically summarized the current deep learning-based methods for isointense infant brain MR image segmentation. We first introduced an authoritative isointense infant brain segmentation dataset, which was used in the iSeg-2017 challenge, hosted as a part of the Medical Image Computing and Computer Assisted Intervention Society Conference 2017. Afterward, several evaluation metrics, including Dice coefficient, 95th-percentile Hausdorff distance, and average symmetric surface distance, were briefly described. We classified the existing deep learning-based methods for isointense infant brain MR image segmentation into two categories: 2D and fully convolutional neural network-based methods. Fully convolutional neural network-based methods can be further divided into two subcategories: 2D and 3D network-based approaches. On the basis of the two categories, we comprehensively described and analyzed the basic ideas, network architecture, improvement schemes, and segmentation performance of each method. In addition, we compared the performance of some existing deep learning-based methods and summarized the analysis results of typical methods on the iSeg-2017 dataset. Lastly, three possible research directions of isointense infant brain MR image segmentation methods based on deep learning were discussed. We drew three main conclusions by reviewing the main work in this field, thereby providing a good overview of existing deep learning methods for isointense infant brain MR image segmentation. First, using multimodality data is beneficial for the network to obtain rich feature information, which can improve the segmentation accuracy. Second, compared with 2D fully convolutional network-based methods, 3D-based methods for MR image brain segmentation can integrate richer spatial context information, resulting in higher accuracy and efficiency. Third, adopting complicated network architecture and dense network connection could make the network achieve superior accuracy performance. Three possible future directions, namely, embedding powerful feature representation modules (e.g., attention mechanism), adding prior knowledge of the infant brain to the network model (e.g., the cortical thickness of infant brain is within a certain range), and constructing a semisupervised or weakly supervised network model trained with a small amount of labeled data, were recommended. Precise segmentation of infant brain tissues is an essential step towards comprehensive volumetric studies and quantitative analysis of early brain development. Deep learning has shown great advantages in isointense infant brain segmentation, and its accuracy and efficiency have been greatly improved compared with traditional methods. With the development of deep learning, it will bring further improvement in the research field of isointense infant brain segmentation and promote the research of early human brain development.关键词:magnetic resonance imaging;isointense phase;infant brain segmentation;deep learning;convolutional neural network101|135|4更新时间:2024-05-07 -
 摘要:Residual neural network (ResNet) has gained considerable attention in deep learning research over the last few years and has made great achievements in computer vision. The deep convolutional network represented by ResNet is increasingly used in the field of medical imaging and has achieved good results in the clinical diagnosis, staging, metastasis, treatment decision, and target area delineation of major diseases, such as tumors, cardiovascular and cerebrovascular diseases, and nervous system diseases. The optimization of the ResNet algorithm is an important part of the ResNet research. It largely determines model performance, such as generalization and convergence. This article summarizes the learning optimization of ResNet. First, the optimization of the learning algorithm of ResNet is elaborated, and the six aspects of activation function, loss function, parameter optimization algorithm, learning decay rate algorithm, normalization, and regularization are summarized. Nine improvement methods exist for the activation function; they are sigmoid, tanh, ReLU, PReLU, randomized ReLU, exponential linear units(ELU), softplus function, noisy softplus function, and maxout. The loss function includes 12 types: cross-entropy, mean square, Euclidean distance, contrast, hinge, softmax, L-softmax, A-softmax, L2 softmax, cosine, center, and focus losses. Eight learning rate decay methods, namely, piecewise constant, polynomial, exponential, inverse time, natural exponential, cosine, linear cosine, and noise linear cosine, are summarized. The normalization algorithms include batch normalization and renormalization. The regularization technologies include seven types: input data, data enhancement, early stop method, L1 regularization, L2 regularization, dropout, and dropout connect. Second, the application study of the residual network model in the diagnosis of medical imaging diseases is reviewed. ResNet is used to diagnose six types of diseases: lung tumor, skin disease, breast cancer, brain disease, diabetes, and hematological disease. 1) Lung cancer. Considerable data show that the incidence of lung cancer is increasing yearly, which is a serious threat to human health. Early diagnosis and detection are essential for the treatment of lung cancer. The main contributions of ResNet in lung tumor research are particle swarm optimization(PSo)+convolutional neural network(CNN), intermediate dense projection method+DenseNet, DenseNet+fully convolutional network(FCN), attention mechanism+ResNet, dense network+U-Net, and 3D+CNN. 2) Skin cancer. Malignant melanoma is one of the most common and deadly skin cancers. Melanoma can be cured if it is properly treated in the early stage. The main contributions of ResNet in the diagnosis and research of skin diseases are integrated learning+CNN, multichannel ResNet, ResNet+support vector machine(SVM), integrated learning+ResNet, and whale optimization algorithm+CNN. 3) Breast cancer. It is a malignant tumor in women, which seriously affects their physical and mental health. The main contributions of ResNet in the diagnosis and research of breast cancer include transfer learning+ResNet, decision tree+ResNet, CNN+SVM, DesneNet-Ⅱ, and SE-attention+CNN. 4) Alzheimer's disease (AD). It is an irreversible brain disease accompanied with progressive impairment of memory and cognitive functions. No effective cure method exists for AD. Early diagnosis of AD is particularly important for patient care and treatment. The main contributions of ResNet in the diagnosis and research of brain diseases are DenseNet, multiscale ResNet, 3DResNet, and multitask CNN+3D DenseNet. 5) Diabetic retinopathy (glycemic retinopathy). It is an eye disease induced by long-term diabetes, which will cause the patient to lose sight and eventually lead to blindness. The main contributions of ResNet in relevant diagnosis and research are migration learning+CNN, FCN+ResNet, multicategory ResNet, and ResNet+SVM. 6) Blood diseases. The proportion of white blood cells in liquid is usually an indicator of diseases. The classification and counting of white blood cells are used in the process of diagnosing diseases. White blood cell test plays a vital role in the detection and treatment of leukemia, anemia, and other diseases. The main contributions of ResNet in the diagnosis and research of blood diseases are as follows: deep supervision ResNet, fine-tuned ResNet, deep cross ResNet, and deep supervision FCN. Lastly, the future development of deep learning in medical imaging is summarized. In this paper, the algorithms of ResNet are systematically summarized, which has a positive significance to the research and development of ResNet.关键词:deep learning;residual neural network(ResNet);optimization algorithm;medical image;disease diagnosis151|247|2更新时间:2024-05-07
摘要:Residual neural network (ResNet) has gained considerable attention in deep learning research over the last few years and has made great achievements in computer vision. The deep convolutional network represented by ResNet is increasingly used in the field of medical imaging and has achieved good results in the clinical diagnosis, staging, metastasis, treatment decision, and target area delineation of major diseases, such as tumors, cardiovascular and cerebrovascular diseases, and nervous system diseases. The optimization of the ResNet algorithm is an important part of the ResNet research. It largely determines model performance, such as generalization and convergence. This article summarizes the learning optimization of ResNet. First, the optimization of the learning algorithm of ResNet is elaborated, and the six aspects of activation function, loss function, parameter optimization algorithm, learning decay rate algorithm, normalization, and regularization are summarized. Nine improvement methods exist for the activation function; they are sigmoid, tanh, ReLU, PReLU, randomized ReLU, exponential linear units(ELU), softplus function, noisy softplus function, and maxout. The loss function includes 12 types: cross-entropy, mean square, Euclidean distance, contrast, hinge, softmax, L-softmax, A-softmax, L2 softmax, cosine, center, and focus losses. Eight learning rate decay methods, namely, piecewise constant, polynomial, exponential, inverse time, natural exponential, cosine, linear cosine, and noise linear cosine, are summarized. The normalization algorithms include batch normalization and renormalization. The regularization technologies include seven types: input data, data enhancement, early stop method, L1 regularization, L2 regularization, dropout, and dropout connect. Second, the application study of the residual network model in the diagnosis of medical imaging diseases is reviewed. ResNet is used to diagnose six types of diseases: lung tumor, skin disease, breast cancer, brain disease, diabetes, and hematological disease. 1) Lung cancer. Considerable data show that the incidence of lung cancer is increasing yearly, which is a serious threat to human health. Early diagnosis and detection are essential for the treatment of lung cancer. The main contributions of ResNet in lung tumor research are particle swarm optimization(PSo)+convolutional neural network(CNN), intermediate dense projection method+DenseNet, DenseNet+fully convolutional network(FCN), attention mechanism+ResNet, dense network+U-Net, and 3D+CNN. 2) Skin cancer. Malignant melanoma is one of the most common and deadly skin cancers. Melanoma can be cured if it is properly treated in the early stage. The main contributions of ResNet in the diagnosis and research of skin diseases are integrated learning+CNN, multichannel ResNet, ResNet+support vector machine(SVM), integrated learning+ResNet, and whale optimization algorithm+CNN. 3) Breast cancer. It is a malignant tumor in women, which seriously affects their physical and mental health. The main contributions of ResNet in the diagnosis and research of breast cancer include transfer learning+ResNet, decision tree+ResNet, CNN+SVM, DesneNet-Ⅱ, and SE-attention+CNN. 4) Alzheimer's disease (AD). It is an irreversible brain disease accompanied with progressive impairment of memory and cognitive functions. No effective cure method exists for AD. Early diagnosis of AD is particularly important for patient care and treatment. The main contributions of ResNet in the diagnosis and research of brain diseases are DenseNet, multiscale ResNet, 3DResNet, and multitask CNN+3D DenseNet. 5) Diabetic retinopathy (glycemic retinopathy). It is an eye disease induced by long-term diabetes, which will cause the patient to lose sight and eventually lead to blindness. The main contributions of ResNet in relevant diagnosis and research are migration learning+CNN, FCN+ResNet, multicategory ResNet, and ResNet+SVM. 6) Blood diseases. The proportion of white blood cells in liquid is usually an indicator of diseases. The classification and counting of white blood cells are used in the process of diagnosing diseases. White blood cell test plays a vital role in the detection and treatment of leukemia, anemia, and other diseases. The main contributions of ResNet in the diagnosis and research of blood diseases are as follows: deep supervision ResNet, fine-tuned ResNet, deep cross ResNet, and deep supervision FCN. Lastly, the future development of deep learning in medical imaging is summarized. In this paper, the algorithms of ResNet are systematically summarized, which has a positive significance to the research and development of ResNet.关键词:deep learning;residual neural network(ResNet);optimization algorithm;medical image;disease diagnosis151|247|2更新时间:2024-05-07 - 摘要:Cervical cancer is an important public health problem worldwide, and its high incidence and trends in younger generation also attract increasing attention. The biological behavior and prognosis of cervical cancer are closely related to its histological type and lymph node metastasis. Cervical cancer has many histological types: the most common type is squamous cell carcinoma, accounting for about 80%; the second most common type is adenocarcinoma, accounting for about 15%~20%, whose incidence is increasing in recent years because of the prevalence of cervical cancer screening. The rare types are adenosquamous carcinoma and neuroendocrine tumors (small cell carcinoma). Their incidence is about 5% and is increasing in recent years because of the cervical cancer screening. Cervical squamous cell carcinoma is more sensitive to radiotherapy and has a better prognosis, whereas cervical adenocarcinoma is less sensitive to radiotherapy and prone to lymph node and hematogenous metastasis. Small cell carcinomas are sensitive to chemotherapies due to their particular origin, prone to early lymph node metastasis, and have poor prognosis. Determining lymphatic metastasis based on magnetic resonance imaging (MRI) is the main factor to evaluate the prognosis of cervical cancer and formulate treatment plan. In the early diagnosis of cervical cancer, 10%~30% of patients have lymph node metastasis. Thus, the evaluation of pelvic and retroperitoneal lymph node metastasis of cervical cancer before treatment will be directly related to the choice of treatment options and prognosis of patients. Although cervical conization and lymph node biopsy are the gold standard for the diagnosis of cervical cancer histopathology and metastatic lymph nodes, the limitations of sampling and the heterogeneity of tumors do not fully reflect the histological information of primary lesions and metastatic lymph nodes of cervical cancer, and biopsy can increase the risk of tumor dissemination.MRI is the commonly used imaging diagnosis and staging method for cervical cancer before surgery. Intravoxel incoherent motion diffusion-weighted imaging (IVIM-DWI), a new functional imaging technique for MRI, is a multi-b-value and bi-exponential model diffusion-weighted technique, which can separate the random diffusion motion of water molecules from the microvascular perfusion effect and reflect the diffusion information of biological tissues. The main parameters are as follows: ADC, D, D*, and f value. The ADCstand value is similar to the ADC(apparent diffusion coefficient) value in a single-value DWI(diffusion weighted imaging) model, which reflects the combined effect of tissue diffusion and perfusion but can simply reflect the diffusion movement of water molecules in tissues. Given the influence of the elevation of microcirculation blood perfusion, the measured values tend to be high. The D value reflects Brownian motion information of pure water molecules in tissues and indirectly reflects cell density in tumors. The D* value and f value provide information of microcirculatory blood flow perfusion in tumors. Moreover, the D* value primarily reflects the blood velocity of microcirculation, whereas the f value primarily reflects the blood volume of microcirculation. Therefore, IVIM-DWI based on multiple parameters can play an important role in tumor characterization, staging, typing, and prediction of lymph node metastasis. At present, artificial intelligence (AI) plays a role in all aspects of people's life. Texture analysis (TA) technology is an important branch of AI research. Medically, TA has become a new research hotspot in Radiomics. TA uses corresponding computer software to extract texture features of lesion tissues from images for objective and quantitative analysis based on imaging images, which can detect microscopic changes of tumor tissues that cannot be recognized by the human eyes, and reveal gray distribution and quantitative data features in tumor tissues. To date, TA is based on images such as computed tomography(CT), MRI, or positron emission tomography(PET)/CT. Three methods are identified to obtain texture features: statistics based, transformation based, and structure based. Among the methods, the most commonly used method is statistics based. Texture features include first-order features, second-order features, and high-order features. First-order features, also known as histogram analysis, describe the gray distribution of individual pixel values in the region of interest, including mean, variance, skewness, kurtosis, and entropy. Second-order features represent local texture features on the basis of the relationship between adjacent 2 pixels. Common methods include gray-level co-occurrence matrix, gray-level run-length matrix, and gray-level area-size matrix. In addition, high-order features analyze local image information by applying gray-level difference matrix of adjacent pixels to reflect the change of local intensity or the distribution of homogeneous regions. TA can quantify the distribution of texture features, such as signal intensity distribution, morphology, and heterogeneity at the lesion site, to reflect the lesion features objectively and comprehensively. Therefore, TA plays an important role in the qualitative, definitive, and differential diagnosis of diseases. Researchers interpret texture information in images on the basis of imaging images, combined with imaging technology and computer AI technology, through the analysis of quantitative data, which provides a possibility for clinical preoperative prediction of different histological types of cervical cancer and metastatic lymph nodes. This article reviews the recent research progress in the clinical application of IVIM-DWI and TA in the preoperative identification of histological subtypes and lymph node metastasis of cervical cancer.关键词:cervical cancer;intravoxel incoherent motion diffusion-weighted imaging(IVIM-DWI);texture analysis(TA);lymph node;metastasis47|79|1更新时间:2024-05-07
- 摘要:Clinicians and experimenters can obtain a time series of three-dimensional images in a fixed time period, that is, 4D (3D + time) longitudinal data with both time and space information, such as two-photon microscopy, CT/MRI(computer tomography/magnetic resonance imaging) Cine scanning mode, or artificially select a time point to scan, and use 4D tensor representation to integrate the collected data in one time and three spatial dimensions, which is suitable for longitudinal analysis. Longitudinal analysis describes the collection of data on one or more variables of the same object at multiple time points to study their changes over time or a set of diachronic research methods that track the influence of some variables, which are commonly used in the medical field: disease changes and causes. However, many studies in the past sliced 4D data into 2D/3D pictures due to the lack of suitable processing algorithms, resulting in significant information loss. In recent years, artificial intelligence has given full play to its natural advantages in massive data processing and has brought new solutions to solve the problems of 4D longitudinal data with high dimensions, large amount of calculation, and difficulty in analysis. Among the solutions, convolutional neural networks, long- and short-term memory networks, and other deep learning algorithms have achieved good results in the processing of different modalities, such as natural language, audio, image, and video. They have exceeded the traditional methods. In the longitudinal medical image analysis using 4D data, deep learning uses spatial information and time-varying information and plays an important role in studying the dynamic changes of goals over time. The main application directions can be divided into two categories: moving target tracking and positioning in the field of biomedicine and tumor growth prediction and auxiliary diagnosis, where moving target tracking and positioning includes matching between complex moving targets in the biological field, 4D longitudinal medical imaging, automatic data segmentation, and vascular dynamics research. Tumor growth prediction and auxiliary diagnosis use volume longitudinal data with time information to model tumor growth, calculate the change in tumor size over a period of time, that is, the growth characteristics of the tumor, and assist the doctor in the diagnostic stage from the perspective of growth rate (benign tumors generally grow slower than malignant tumors). Cancer grades are recommended for patients to review. During the treatment phase, tumor changes are accurately measured; the effects of radiotherapy or drug treatment are evaluated; survival is predicted; personalized treatment plans for patients are developed, and drug development is promoted. However, the abovementioned longitudinal analysis only focuses on the change of the macroscopic morphology of the lesion and cannot reflect all tumor biological information or predict the clinically relevant tumor properties. In the future, horizontal correlation analysis can include temporal features, capture the relationship between temporal and spatial changes, and map the quantitative values of molecular features to histopathological changes, thereby proving the relationship between the secretion of cytokines and the severity of lesions in the image relevance to explain the causality of the disease from the table and achieve personalized treatment. In addition, the accuracy and generalization of the abovementioned artificial intelligence algorithms depend on large-scale and high-quality data. Medical data have fewer samples, larger acquisition costs, and higher resolution requirements than data in other fields. Problem and longitudinal data make data collection difficult because they contain time information not previously involved. Future medical longitudinal data points can be combined with federal learning and transfer learning: federal learning is used to ensure that no data exchange is observed while improving the generalization of the model, making the same model suitable for data from other hospitals, and protecting the privacy of medical data. On the contrary, transfer learning is used to solve the problem of few high-quality samples and the lack of frame-by-frame labels, improve the accuracy of the model, and solve hidden security risks such as user privacy when the product is launched in the future.关键词:artificial intelligence;medical image processing;longitudinal analysis;4D image;spatiotemporal data152|166|0更新时间:2024-05-07
Frontier
-
 摘要:ObjectiveAccurate delineation of organs at risk (OARs) is an essential step in the radiation therapy for cancers. However, this procedure is frequently time consuming and error prone because of the large anatomical variation across patients, different experience of observers, and poor soft-tissue contrast in computed tomography (CT) scans. A computer-aided analysis system for OAR auto-segmentation from CT images will reduce the burden of doctors and the subjective errors and improve the effect of radiotherapy. In the early years, atlas-based methods are extremely popular and widely used in anatomy segmentation. However, the performance of atlas-based segmentation methods can be easily affected by various factors, such as the quality of atlas and registration methods. Recently, profits from the rapid growth of computing power and the amount of available data, deep learning, especially deep convolutional neural networks (CNNs), has shown great potential in the field of image analysis. For most of the medical image segmentation tasks, the algorithms based on CNN outperform traditional methods. As a special fully CNN, U-Net adopts the design of encoder-decoder and fuses the high- and low-level features by skip connections to realize pixelwise segmentation. Given the outstanding performance of U-Net, numerous derivatives of the U-Net architecture have been gradually developed in various organ segmentation tasks. V-Net is proposed as an improvement scheme of U-Net to address the difficulties in processing 3D data. V-Net can fully utilize the 3D characteristics of images, although it is unsuitable for the 3D medical image datasets with few samples and the segmentation tasks of small organs. Therefore, a two-step 2D CNN model is proposed for the automatic and accurate segmentation of OARs in radiotherapy.MethodIn this study, we propose a novel cascade-CNN model that mainly includes a slice classifier and a 2D organ segmentation network. visual geometry group (VGG)16 is used as the backbone structure of the classifier and modified accordingly by considering the time overhead and the size of available data. To reduce the parameters and calculation complexity, three convolutional layers are removed, and an additional global max pooling layer is used as the bridging module between the feature extraction and the fully connected layer. The segmentation network is built upon the U-Net by replacing the deconvolutional layers with bilinear interpolation layers to perform the upsampling for features. The dropout layer and the data augmentation are utilized to avoid the overfitting problem. The classifier and the segmentation network for each organ are implemented in Keras and independently trained with the binary cross-entropy and dice losses, respectively. We use adaptive moment estimation to stabilize the gradient descent process during the training of the two models. In the inference stage, the slices containing the target organs are first selected by the classifier from the entire CT scans. These slices are then used as the input of the segmentation network to obtain the results, and some simple post-processing methods are applied to optimize the segmentation results.ResultThe dataset containing CT scans of 89 cases of cervical cancer patients and the manual segmentation results provided by multiple radiologists in The First Affiliated Hospital of University of Science and Technology of China (USTC) are used as the gold standard for evaluation. In the experimental part, the average classification accuracy, precision, recall, and F1-score of the classifier on the six organs (left femoral head, right femoral head, left femur, right femur, bladder, and rectum) are 98.36%, 96.64%, 94.1%, and 95.34%, respectively. On the basis of the performance of the above classifiers, the proposed method achieves high segmentation accuracy on the bladder, left femoral head, right femoral head, left femur, and right femur, with Dice coefficients of 94.16%, 93.69%, 95.09%, 96.14%, and 96.57%, respectively. Compared with the single U-Net and cascaded V-Net, the Dice coefficient increases by 4.1% to 6.6%. For the rectum, all methods perform poorly because of the irregular shape and low contrast. The proposed method achieves a Dice coefficient of 72.99% on the rectum, which is approximately 20% higher than other methods. The comparison experiments demonstrate that the classifiers can effectively improve the overall segmentation accuracy.ConclusionIn this work, we propose a novel 2D cascade-CNN model, which is composed of a classifier and a segmentation network, for the automatic segmentation of OARs in radiation therapy. The experiments demonstrate that the proposed method effectively alleviates the labeling sparse problem and reduces the false positive segmentation results. For the organs that immensely vary in shape and size, the proposed method obtains a significant improvement in segmentation accuracy. In comparison with existing neural network methods, the proposed method achieves state-of-the-art results in the segmentation task of OARs for cervical cancer and a better consistency with that of experienced radiologists.关键词:segmentation of organs at risk;convolutional neural network(CNN);cascade model;radiation therapy;cervical cancer84|198|0更新时间:2024-05-07
摘要:ObjectiveAccurate delineation of organs at risk (OARs) is an essential step in the radiation therapy for cancers. However, this procedure is frequently time consuming and error prone because of the large anatomical variation across patients, different experience of observers, and poor soft-tissue contrast in computed tomography (CT) scans. A computer-aided analysis system for OAR auto-segmentation from CT images will reduce the burden of doctors and the subjective errors and improve the effect of radiotherapy. In the early years, atlas-based methods are extremely popular and widely used in anatomy segmentation. However, the performance of atlas-based segmentation methods can be easily affected by various factors, such as the quality of atlas and registration methods. Recently, profits from the rapid growth of computing power and the amount of available data, deep learning, especially deep convolutional neural networks (CNNs), has shown great potential in the field of image analysis. For most of the medical image segmentation tasks, the algorithms based on CNN outperform traditional methods. As a special fully CNN, U-Net adopts the design of encoder-decoder and fuses the high- and low-level features by skip connections to realize pixelwise segmentation. Given the outstanding performance of U-Net, numerous derivatives of the U-Net architecture have been gradually developed in various organ segmentation tasks. V-Net is proposed as an improvement scheme of U-Net to address the difficulties in processing 3D data. V-Net can fully utilize the 3D characteristics of images, although it is unsuitable for the 3D medical image datasets with few samples and the segmentation tasks of small organs. Therefore, a two-step 2D CNN model is proposed for the automatic and accurate segmentation of OARs in radiotherapy.MethodIn this study, we propose a novel cascade-CNN model that mainly includes a slice classifier and a 2D organ segmentation network. visual geometry group (VGG)16 is used as the backbone structure of the classifier and modified accordingly by considering the time overhead and the size of available data. To reduce the parameters and calculation complexity, three convolutional layers are removed, and an additional global max pooling layer is used as the bridging module between the feature extraction and the fully connected layer. The segmentation network is built upon the U-Net by replacing the deconvolutional layers with bilinear interpolation layers to perform the upsampling for features. The dropout layer and the data augmentation are utilized to avoid the overfitting problem. The classifier and the segmentation network for each organ are implemented in Keras and independently trained with the binary cross-entropy and dice losses, respectively. We use adaptive moment estimation to stabilize the gradient descent process during the training of the two models. In the inference stage, the slices containing the target organs are first selected by the classifier from the entire CT scans. These slices are then used as the input of the segmentation network to obtain the results, and some simple post-processing methods are applied to optimize the segmentation results.ResultThe dataset containing CT scans of 89 cases of cervical cancer patients and the manual segmentation results provided by multiple radiologists in The First Affiliated Hospital of University of Science and Technology of China (USTC) are used as the gold standard for evaluation. In the experimental part, the average classification accuracy, precision, recall, and F1-score of the classifier on the six organs (left femoral head, right femoral head, left femur, right femur, bladder, and rectum) are 98.36%, 96.64%, 94.1%, and 95.34%, respectively. On the basis of the performance of the above classifiers, the proposed method achieves high segmentation accuracy on the bladder, left femoral head, right femoral head, left femur, and right femur, with Dice coefficients of 94.16%, 93.69%, 95.09%, 96.14%, and 96.57%, respectively. Compared with the single U-Net and cascaded V-Net, the Dice coefficient increases by 4.1% to 6.6%. For the rectum, all methods perform poorly because of the irregular shape and low contrast. The proposed method achieves a Dice coefficient of 72.99% on the rectum, which is approximately 20% higher than other methods. The comparison experiments demonstrate that the classifiers can effectively improve the overall segmentation accuracy.ConclusionIn this work, we propose a novel 2D cascade-CNN model, which is composed of a classifier and a segmentation network, for the automatic segmentation of OARs in radiation therapy. The experiments demonstrate that the proposed method effectively alleviates the labeling sparse problem and reduces the false positive segmentation results. For the organs that immensely vary in shape and size, the proposed method obtains a significant improvement in segmentation accuracy. In comparison with existing neural network methods, the proposed method achieves state-of-the-art results in the segmentation task of OARs for cervical cancer and a better consistency with that of experienced radiologists.关键词:segmentation of organs at risk;convolutional neural network(CNN);cascade model;radiation therapy;cervical cancer84|198|0更新时间:2024-05-07 -
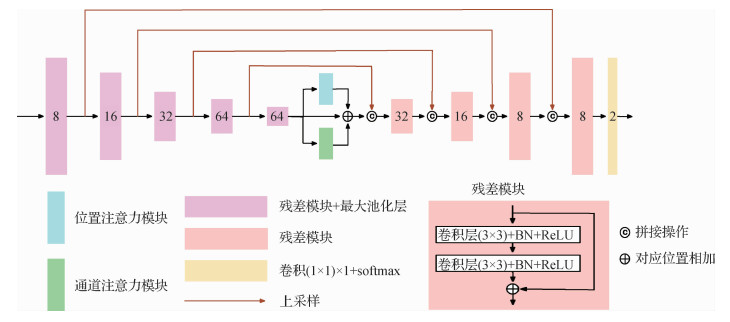 摘要:ObjectivePrecise lung tumor segmentation is a necessary step in computer-aided diagnosis, surgical planning, and radiotherapy of lung cancer. Computed tomography (CT) images are important auxiliary tools in clinical medicine. The diagnosis of lung cancer tumors is labor intensive that requires professional radiologists to carefully examine hundreds of CT slices for finding and confirming the location of tumor lesions, and final reports need to be verified by other experienced radiologists. This process consumes time and effort. Doctors commonly make different diagnoses at the same time, and the same doctor may make different decisions at different times because of the difference in their subjective experience. To solve the above problems, increasing scientific researchers have devoted to the field of medical imaging by continuously promoting the combination of artificial intelligence and medical imaging, and the automatic segmentation of lung tumors has been widely investigated. To address the problems that 3D U-Net is insufficiently accurate and is prone to produce false positive pixels, this paper proposes a new network named dual attention U-Net (DAU-Net) that incorporates dual attention mechanisms and residual modules. A post processing method based on connected component analysis is used to remove the false positive regions outside the region of interest.MethodIn accordance with the characteristics of lung CT images, we proposed a pipeline to preprocess CT images, which was divided into three steps. Standardizing pixel pitch was the first step that needs to be performed because different pixel spacings will affect the speed and quality of network convergence in the training process. The thickness of all 2D slices is 5 mm, and the range of in-plane resolution varies from 0.607 mm to 0.976 mm. Thus, linear interpolation was applied to each CT slice to obtain 1 mm in-plane resolution. The interpolated CT images still exist in 3D form. The window width and window level were then set to 1 600 and -200, respectively, that is, the pixel values in the CT image greater than 600 were set to 600 and those less than -1 000 were set to -1 000. The intensity values of images were truncated to the range of [-1 000, 600] and linearly normalized to [0, 1] to enhance the regions of interest when using CT images, which is helpful for the automatic segmentation of lesions. This step will make the size of each CT image less than N×512×512, where N is the number of slices. After padding to N×512×512, the CT images and their corresponding annotations were cropped to a constant size of N×320×260 from a fixed coordinate (0, 90, 130) of the very beginning slice, and interpolation was used to scale the size of the images to 64×320×260. The main architecture of the network adopts the 3D form of the U-Net by replacing every two adjacent convolutional layers with a residual structure and adding two attention mechanisms to the middle of the contraction path and the expansion path to obtain DAU-Net. The network can alleviate degradation, gradient disappearance, and gradient explosion caused by the increase in the depth of the neural network by adding the residual structures. Similar to U-Net, encoder-decoder networks can merge high-resolution feature maps with position information and low-resolution feature maps with contextual information through skip connections to capture targets of different scales. However, they cannot take advantage of the positional relationship of different objects in global images and association between different categories. To retain the advantages of encoder-decoder structures and overcome the above problems, a position attention module and a channel attention module connected in parallel are combined with 3D U-Net. The position attention module can encode context information from a wide range into local features and the channel attention module can find the dependency relationship between different channels, thereby strengthening the interdependent features. The network can perform end-to-end training and it was trained by optimizing soft dice loss in this work. After inference, connected component analysis is used to remove the false positive regions that are wrongly segmented by only keeping the largest connected component and discarding other parts. Considering that this paper uses a 3D CNN(convolutional neural network), a 26-neighborhood connected component analysis method is used to determine the connection relationship between a central pixel and its 26 adjacent pixels. The output of the network has two channels, and softmax is used to make the output between zero and one. In binarization, only the channel index with a high probability is selected to obtain the final binary result where the connected component analysis method is applied. This postprocessing method effectively improves the segmentation accuracy and decreases the false positive rate (FPR). The premise of using this method is that the dataset we use contains only one lesion per case.ResultWe retrospectively collected data from patients in Shanghai Chest Hospital from 2013 to 2017. The study was approved by Shanghai Chest Hospital, Shanghai Jiao Tong University. Ethical approval (ID: KS 1716) was obtained for use of the CT images. Experienced radiologists provided the gold standard of each case. In the experiment, we compared the standard 3D U-Net and the reproduced 3D attention U-Net. The experiment used 10-fold cross-validation for all networks, and we adopted the widely used Dice, Hausdorff distance (HD), FPR, and true positive rate to evaluate the predicted outputs. The results show that the proposed DAU-Net has powerful performance in the lung tumor segmentation task, and the postprocessing method can effectively reduce the interference of false positive regions on the segmentation results. Compared with 3D U-Net, Dice and HD are improved by 2.5% and 9.7%, respectively, and FPR is reduced by 13.6%.ConclusionThe proposed lung tumor segmentation algorithm can effectively improve the accuracy of tumor segmentation and help to achieve rapid, stable, and accurate segmentation of lung cancer.关键词:U-Net;computed tomography(CT);lung tumor;segmentation;attention mechanism144|78|8更新时间:2024-05-07
摘要:ObjectivePrecise lung tumor segmentation is a necessary step in computer-aided diagnosis, surgical planning, and radiotherapy of lung cancer. Computed tomography (CT) images are important auxiliary tools in clinical medicine. The diagnosis of lung cancer tumors is labor intensive that requires professional radiologists to carefully examine hundreds of CT slices for finding and confirming the location of tumor lesions, and final reports need to be verified by other experienced radiologists. This process consumes time and effort. Doctors commonly make different diagnoses at the same time, and the same doctor may make different decisions at different times because of the difference in their subjective experience. To solve the above problems, increasing scientific researchers have devoted to the field of medical imaging by continuously promoting the combination of artificial intelligence and medical imaging, and the automatic segmentation of lung tumors has been widely investigated. To address the problems that 3D U-Net is insufficiently accurate and is prone to produce false positive pixels, this paper proposes a new network named dual attention U-Net (DAU-Net) that incorporates dual attention mechanisms and residual modules. A post processing method based on connected component analysis is used to remove the false positive regions outside the region of interest.MethodIn accordance with the characteristics of lung CT images, we proposed a pipeline to preprocess CT images, which was divided into three steps. Standardizing pixel pitch was the first step that needs to be performed because different pixel spacings will affect the speed and quality of network convergence in the training process. The thickness of all 2D slices is 5 mm, and the range of in-plane resolution varies from 0.607 mm to 0.976 mm. Thus, linear interpolation was applied to each CT slice to obtain 1 mm in-plane resolution. The interpolated CT images still exist in 3D form. The window width and window level were then set to 1 600 and -200, respectively, that is, the pixel values in the CT image greater than 600 were set to 600 and those less than -1 000 were set to -1 000. The intensity values of images were truncated to the range of [-1 000, 600] and linearly normalized to [0, 1] to enhance the regions of interest when using CT images, which is helpful for the automatic segmentation of lesions. This step will make the size of each CT image less than N×512×512, where N is the number of slices. After padding to N×512×512, the CT images and their corresponding annotations were cropped to a constant size of N×320×260 from a fixed coordinate (0, 90, 130) of the very beginning slice, and interpolation was used to scale the size of the images to 64×320×260. The main architecture of the network adopts the 3D form of the U-Net by replacing every two adjacent convolutional layers with a residual structure and adding two attention mechanisms to the middle of the contraction path and the expansion path to obtain DAU-Net. The network can alleviate degradation, gradient disappearance, and gradient explosion caused by the increase in the depth of the neural network by adding the residual structures. Similar to U-Net, encoder-decoder networks can merge high-resolution feature maps with position information and low-resolution feature maps with contextual information through skip connections to capture targets of different scales. However, they cannot take advantage of the positional relationship of different objects in global images and association between different categories. To retain the advantages of encoder-decoder structures and overcome the above problems, a position attention module and a channel attention module connected in parallel are combined with 3D U-Net. The position attention module can encode context information from a wide range into local features and the channel attention module can find the dependency relationship between different channels, thereby strengthening the interdependent features. The network can perform end-to-end training and it was trained by optimizing soft dice loss in this work. After inference, connected component analysis is used to remove the false positive regions that are wrongly segmented by only keeping the largest connected component and discarding other parts. Considering that this paper uses a 3D CNN(convolutional neural network), a 26-neighborhood connected component analysis method is used to determine the connection relationship between a central pixel and its 26 adjacent pixels. The output of the network has two channels, and softmax is used to make the output between zero and one. In binarization, only the channel index with a high probability is selected to obtain the final binary result where the connected component analysis method is applied. This postprocessing method effectively improves the segmentation accuracy and decreases the false positive rate (FPR). The premise of using this method is that the dataset we use contains only one lesion per case.ResultWe retrospectively collected data from patients in Shanghai Chest Hospital from 2013 to 2017. The study was approved by Shanghai Chest Hospital, Shanghai Jiao Tong University. Ethical approval (ID: KS 1716) was obtained for use of the CT images. Experienced radiologists provided the gold standard of each case. In the experiment, we compared the standard 3D U-Net and the reproduced 3D attention U-Net. The experiment used 10-fold cross-validation for all networks, and we adopted the widely used Dice, Hausdorff distance (HD), FPR, and true positive rate to evaluate the predicted outputs. The results show that the proposed DAU-Net has powerful performance in the lung tumor segmentation task, and the postprocessing method can effectively reduce the interference of false positive regions on the segmentation results. Compared with 3D U-Net, Dice and HD are improved by 2.5% and 9.7%, respectively, and FPR is reduced by 13.6%.ConclusionThe proposed lung tumor segmentation algorithm can effectively improve the accuracy of tumor segmentation and help to achieve rapid, stable, and accurate segmentation of lung cancer.关键词:U-Net;computed tomography(CT);lung tumor;segmentation;attention mechanism144|78|8更新时间:2024-05-07 -
 摘要:ObjectiveIn clinical diagnosis,the manual segmentation of liver tumor needs to have anatomical knowledge and experience,which is highly subjective. Automatic segmentation of liver tumor based on computed tomography(CT) medical image research is important in guiding clinical diagnosis and treatment. It can form accurate preoperative prediction,intraoperative monitoring,and postoperative evaluation,which develop a perfect surgical treatment plan and improve the success rate of liver tumor surgery. However,a typical medical image is complex,which may contain many organs and tissues. In the imaging process,it often produces more or less interference information due to the influence of imaging equipment and human factors. According to the imaging principle and characteristics of medical imaging,not all information may be helpful for medical diagnosis. Whether CT or magnetic resonance imaging(MRI),as the common chest and abdomen scanning,in the imaging effect,the image contrast is low; the boundary is fuzzy; most of the time,accurate segmentation of interested organs and corresponding tumors requires clinical anatomy knowledge of doctors. In general,many boundaries are achieved through consultation between medical personnel. Notably,the automatic and accurate segmentation of liver tumor in medical images is a great challenge. In this paper,an effective segmentation method is proposed by combining multiple deep learning techniques with radiomics.MethodFirst,a cascaded 2D image end-to-end segmentation model is established to segment the liver and tumor simultaneously. The segmentation model adopts the U-Net deep network framework,which connects the internal modules of the encoder and decoder and the layers of the encoder and decoder intensively. This diversified feature fusion can obtain accurate global position features and abundant local details or physical characteristics. Sub-pixel convolution is added during upsampling by pixel shuffling multiple low-resolution feature images to output the high-resolution image and make up for the loss of some image texture details. Given the small area and volume of the small tumor area,feature disappearance or weakening can easily occur in the convolutional neural network. A new attention gate model for medical imaging is used here to improve the segmentation performance of the tumor. This model can automatically learn the target with different volumes and shapes,which is suitable for liver tumor with the distribution characteristics of volume and shape polymorphism. Morphological expansion and corrosion may lead to the loss of some small areas,which are not continuous. Considering that CT scan is continuous in a short time,the lost areas can be recovered by using the continuity of this image in time. If there are segmentation regions in the former and latter image,but the current image does not,then the average value of the segmentation results before and after the current image is used. Given the heterogeneity of malignant tumors,the radiomic characteristics can be an effective,non-invasive imaging biomarker. In this paper,the radiomic features are divided into eight groups: histogram statistical features,gradient features,run-length texture features,gray-level co-occurrence matrix texture features,shape-based features,second-order moment features,boundary features,and three-dimensional wavelet features. According to a known research scope,considerable domestic and foreign research literature of radiomics show that the comprehensive performance of the random forest model is the best,and the random forest classifier is also selected to remove the false-positive tumor. Some segmentation areas are more accurate after morphological operation,but some segmentation areas will be larger than the real results. Therefore,the edge contour must be further refined. Here,the classification based on 3D image blocks does not need complex convolutional neural network(CNN),and simple CNN can achieve a good classification effect. The AlexNet model framework is selected and adjusted accordingly to refine the contour.ResultThe experimental data are from 300 cases of liver cancer in the imaging department of the cooperative hospital. The liver and tumor in each sequence are segmented and labeled by medical experts for more than 10 years. The average accuracy of sensitivity,positive predicted value,and Dice coefficient in the validation results are 0.87 ±0.03,0.91 ±0.03,and 0.86 ±0.05,respectively,which are improved by 0.03,0.02,and 0.04,respectively,compared with the model with the second performance. The training process of the method proposed in this paper is not end-to-end but is completed by three learning models,which are generally carried out on the GPU server under the Linux system. After the model training,the image can be arranged in the server or the general PC client. If the image is in the server,the image needs to request and transmit the sequence image through the network and then return to the client after segmentation. A typical way is to upload the image to the server and perform offline batch segmentation. After processing,only the segmentation result data will be returned.ConclusionIn clinical diagnosis,the automatic or semi-automatic segmentation of the liver and tumor in CT medical image by using artificial intelligence and computer vision technology gradually replaces the pure artificial operation mode. The method proposed in this paper is not limited to the segmentation of liver tumors but also applicable to the segmentation of other medical imaging organs and tumors. In the future research,we will integrate the generative adversarial network technology to improve the segmentation performance.关键词:deep learning;radiomics;full convolution network;attention model;dense connection107|251|6更新时间:2024-05-07
摘要:ObjectiveIn clinical diagnosis,the manual segmentation of liver tumor needs to have anatomical knowledge and experience,which is highly subjective. Automatic segmentation of liver tumor based on computed tomography(CT) medical image research is important in guiding clinical diagnosis and treatment. It can form accurate preoperative prediction,intraoperative monitoring,and postoperative evaluation,which develop a perfect surgical treatment plan and improve the success rate of liver tumor surgery. However,a typical medical image is complex,which may contain many organs and tissues. In the imaging process,it often produces more or less interference information due to the influence of imaging equipment and human factors. According to the imaging principle and characteristics of medical imaging,not all information may be helpful for medical diagnosis. Whether CT or magnetic resonance imaging(MRI),as the common chest and abdomen scanning,in the imaging effect,the image contrast is low; the boundary is fuzzy; most of the time,accurate segmentation of interested organs and corresponding tumors requires clinical anatomy knowledge of doctors. In general,many boundaries are achieved through consultation between medical personnel. Notably,the automatic and accurate segmentation of liver tumor in medical images is a great challenge. In this paper,an effective segmentation method is proposed by combining multiple deep learning techniques with radiomics.MethodFirst,a cascaded 2D image end-to-end segmentation model is established to segment the liver and tumor simultaneously. The segmentation model adopts the U-Net deep network framework,which connects the internal modules of the encoder and decoder and the layers of the encoder and decoder intensively. This diversified feature fusion can obtain accurate global position features and abundant local details or physical characteristics. Sub-pixel convolution is added during upsampling by pixel shuffling multiple low-resolution feature images to output the high-resolution image and make up for the loss of some image texture details. Given the small area and volume of the small tumor area,feature disappearance or weakening can easily occur in the convolutional neural network. A new attention gate model for medical imaging is used here to improve the segmentation performance of the tumor. This model can automatically learn the target with different volumes and shapes,which is suitable for liver tumor with the distribution characteristics of volume and shape polymorphism. Morphological expansion and corrosion may lead to the loss of some small areas,which are not continuous. Considering that CT scan is continuous in a short time,the lost areas can be recovered by using the continuity of this image in time. If there are segmentation regions in the former and latter image,but the current image does not,then the average value of the segmentation results before and after the current image is used. Given the heterogeneity of malignant tumors,the radiomic characteristics can be an effective,non-invasive imaging biomarker. In this paper,the radiomic features are divided into eight groups: histogram statistical features,gradient features,run-length texture features,gray-level co-occurrence matrix texture features,shape-based features,second-order moment features,boundary features,and three-dimensional wavelet features. According to a known research scope,considerable domestic and foreign research literature of radiomics show that the comprehensive performance of the random forest model is the best,and the random forest classifier is also selected to remove the false-positive tumor. Some segmentation areas are more accurate after morphological operation,but some segmentation areas will be larger than the real results. Therefore,the edge contour must be further refined. Here,the classification based on 3D image blocks does not need complex convolutional neural network(CNN),and simple CNN can achieve a good classification effect. The AlexNet model framework is selected and adjusted accordingly to refine the contour.ResultThe experimental data are from 300 cases of liver cancer in the imaging department of the cooperative hospital. The liver and tumor in each sequence are segmented and labeled by medical experts for more than 10 years. The average accuracy of sensitivity,positive predicted value,and Dice coefficient in the validation results are 0.87 ±0.03,0.91 ±0.03,and 0.86 ±0.05,respectively,which are improved by 0.03,0.02,and 0.04,respectively,compared with the model with the second performance. The training process of the method proposed in this paper is not end-to-end but is completed by three learning models,which are generally carried out on the GPU server under the Linux system. After the model training,the image can be arranged in the server or the general PC client. If the image is in the server,the image needs to request and transmit the sequence image through the network and then return to the client after segmentation. A typical way is to upload the image to the server and perform offline batch segmentation. After processing,only the segmentation result data will be returned.ConclusionIn clinical diagnosis,the automatic or semi-automatic segmentation of the liver and tumor in CT medical image by using artificial intelligence and computer vision technology gradually replaces the pure artificial operation mode. The method proposed in this paper is not limited to the segmentation of liver tumors but also applicable to the segmentation of other medical imaging organs and tumors. In the future research,we will integrate the generative adversarial network technology to improve the segmentation performance.关键词:deep learning;radiomics;full convolution network;attention model;dense connection107|251|6更新时间:2024-05-07 -
 摘要:ObjectiveThe Coronavirus Disease 2019 (COVID-19) has become a global pandemic,causing millions of people to be infected worldwide. Imaging analysis based on computed tomography (CT) data is an important means of clinical diagnosis. A supercomputing-supported method is proposed for the construction of a new comprehensive CT analysis auxiliary system dealing with pneumonia.MethodThe system consists of four parts: input processing module,preprocessing module,imaging analysis subsystem,and artificial intelligence(AI) analysis subsystem. Among the four parts,the imaging analysis subsystem detects the pneumonia features,distinguishes typical new coronary pneumonia by analyzing the typical imaging features,such as lung consolidation,ground-glass opacities,and crazy-paving pattern,and then comes to a conclusion of pneumonia. The AI analysis subsystem uses a deep learning model to classify typical viral pneumonia and COVID-19,which enhances the screening ability of pneumonia. Convolutional neural network is widely used as an effective algorithm for medical image analysis,particularly in image classification. It is also widely utilized in the CT image screening and has achieved good results,which has attracted the attention of domestic and foreign scholars and industry. The seasonable result derived from deep learning relies largely on the number and quality of training samples. Given the lack of training samples,the system selects transfer learning as the technical direction for model construction. Considering the quick response to the epidemic,the quality of easy maintenance and dynamic system updating is required. Thus,after comparing and analyzing the performance and classification effect indexes of many common image classification models,we build a transfer learning neural network model on the basis of inception. The entire neural network can be roughly divided into two parts: the first part uses a pre-trained inception network; the role of which is to convert image data into a one-dimensional feature vector. The second part uses a fully connected network to improve classification prediction. The imaging analysis method analyzes the image features of COVID-19,extracts the pneumonia feature areas,and carries out semantic analysis to achieve the delineation of the pneumonia target area. Simultaneously,the typical imaging characteristics of COVID-19 (such as ground glass shadow,infiltration shadow,and lung consolidation) are targeted. With regard to the pneumonia target area,a multi-level dynamic threshold segmentation is first used to determine the minimum lung tissue area (rectangular region of interest (ROI)). The extraction of the lung tissue area is designed as a normal workflow. For each ROI,pixel statistics,threshold segmentation,regional dissolution and expansion,and abnormal proofreading are used to obtain the pneumonia target area. Aiming at the relationship between the sizes of the pneumonia target area,a logical filter is established to detect the segmented distribution features and spatial relationship with the outer contour of the lung. Then,based on the characteristic relationship of typical new coronary pneumonia,the typical characteristics of new coronary pneumonia are outlined. The entire comprehensive analysis platform is built on the basis of the Tianhe artificial intelligence innovation-integrated platform. The Tianhe artificial intelligence innovation-integrated platform is based on the hardware fusion supporting the environment of Tianhe supercomputing,cloud computing,and big data,upon which realizes the existing mainstream deep learning framework. It is highly encapsulated with the processing model algorithm,forming a visual interactive template development environment covering multiple links such as data loading,model construction,training,verification,and solidified deployment. As a service on this supporting platform,CT image comprehensive analysis AI auxiliary system has access to the computing resources,data resources,and external service capabilities of the platform and finally achieves the rapid integration and dynamic update of the system during the pandemic.ResultAfter its release,the system has continuously and steadily provided new COVID-19 auxiliary diagnostic services and scientific research support for more than 30 hospitals and more than 100 scientific research institutions at home and abroad,providing important support for combating the epidemic.ConclusionThe supercomputing-supported new coronary pneumonia CT image comprehensive analysis auxiliary system construction method proposed in this paper has achieved important application on diagnosis and research. It is an effective way to achieve rapid deployment services and provide efficient support for emergencies. The system applies artificial intelligence technology using CT imaging to screen for COVID-19. By applying artificial intelligence to the screening of COVID-19 with pneumonia and giving reference opinions for auxiliary diagnosis,the marking and area statistics of the inflammatory regions are improved. The system achieves the combination of artificial intelligence traditional machine vision and deep learning technology to distinguish COVID-19 by using CT images. The combined route of viral pneumonitis feature extraction based on traditional machine vision and the COVID-19 image classification based on artificial intelligence technology has achieved a comprehensive analysis of medical image features and COVID-19 screening. The fast implementation mode of the fusion platform scenario is based on computing power and data support. Relying on the Tianhe artificial intelligence innovation-integrated service platform,the platform supports intelligent frontier innovation on the basis of computing power and data,implements an open model of simultaneous research and application,and has a multi-industry training resource model library and large-scale distributed training sources. With regard to rapid deployment and other service capabilities,this comprehensive analysis system is also the first public COVID-19 AI-assisted diagnostic system deployed online. Analysis based directly on digital imaging and communications in medicine(DICOM) data and video data will effectively improve the analysis efficiency,but it will involve data ethics and security-related issues; however,it is the developing direction that needs to be resolved in the future.关键词:Coronavirus Disease 2019(COVID-19);computed tomography(CT) imaging;auxiliary diagnosis;artificial intelligence(AI);supercomputing76|156|2更新时间:2024-05-07
摘要:ObjectiveThe Coronavirus Disease 2019 (COVID-19) has become a global pandemic,causing millions of people to be infected worldwide. Imaging analysis based on computed tomography (CT) data is an important means of clinical diagnosis. A supercomputing-supported method is proposed for the construction of a new comprehensive CT analysis auxiliary system dealing with pneumonia.MethodThe system consists of four parts: input processing module,preprocessing module,imaging analysis subsystem,and artificial intelligence(AI) analysis subsystem. Among the four parts,the imaging analysis subsystem detects the pneumonia features,distinguishes typical new coronary pneumonia by analyzing the typical imaging features,such as lung consolidation,ground-glass opacities,and crazy-paving pattern,and then comes to a conclusion of pneumonia. The AI analysis subsystem uses a deep learning model to classify typical viral pneumonia and COVID-19,which enhances the screening ability of pneumonia. Convolutional neural network is widely used as an effective algorithm for medical image analysis,particularly in image classification. It is also widely utilized in the CT image screening and has achieved good results,which has attracted the attention of domestic and foreign scholars and industry. The seasonable result derived from deep learning relies largely on the number and quality of training samples. Given the lack of training samples,the system selects transfer learning as the technical direction for model construction. Considering the quick response to the epidemic,the quality of easy maintenance and dynamic system updating is required. Thus,after comparing and analyzing the performance and classification effect indexes of many common image classification models,we build a transfer learning neural network model on the basis of inception. The entire neural network can be roughly divided into two parts: the first part uses a pre-trained inception network; the role of which is to convert image data into a one-dimensional feature vector. The second part uses a fully connected network to improve classification prediction. The imaging analysis method analyzes the image features of COVID-19,extracts the pneumonia feature areas,and carries out semantic analysis to achieve the delineation of the pneumonia target area. Simultaneously,the typical imaging characteristics of COVID-19 (such as ground glass shadow,infiltration shadow,and lung consolidation) are targeted. With regard to the pneumonia target area,a multi-level dynamic threshold segmentation is first used to determine the minimum lung tissue area (rectangular region of interest (ROI)). The extraction of the lung tissue area is designed as a normal workflow. For each ROI,pixel statistics,threshold segmentation,regional dissolution and expansion,and abnormal proofreading are used to obtain the pneumonia target area. Aiming at the relationship between the sizes of the pneumonia target area,a logical filter is established to detect the segmented distribution features and spatial relationship with the outer contour of the lung. Then,based on the characteristic relationship of typical new coronary pneumonia,the typical characteristics of new coronary pneumonia are outlined. The entire comprehensive analysis platform is built on the basis of the Tianhe artificial intelligence innovation-integrated platform. The Tianhe artificial intelligence innovation-integrated platform is based on the hardware fusion supporting the environment of Tianhe supercomputing,cloud computing,and big data,upon which realizes the existing mainstream deep learning framework. It is highly encapsulated with the processing model algorithm,forming a visual interactive template development environment covering multiple links such as data loading,model construction,training,verification,and solidified deployment. As a service on this supporting platform,CT image comprehensive analysis AI auxiliary system has access to the computing resources,data resources,and external service capabilities of the platform and finally achieves the rapid integration and dynamic update of the system during the pandemic.ResultAfter its release,the system has continuously and steadily provided new COVID-19 auxiliary diagnostic services and scientific research support for more than 30 hospitals and more than 100 scientific research institutions at home and abroad,providing important support for combating the epidemic.ConclusionThe supercomputing-supported new coronary pneumonia CT image comprehensive analysis auxiliary system construction method proposed in this paper has achieved important application on diagnosis and research. It is an effective way to achieve rapid deployment services and provide efficient support for emergencies. The system applies artificial intelligence technology using CT imaging to screen for COVID-19. By applying artificial intelligence to the screening of COVID-19 with pneumonia and giving reference opinions for auxiliary diagnosis,the marking and area statistics of the inflammatory regions are improved. The system achieves the combination of artificial intelligence traditional machine vision and deep learning technology to distinguish COVID-19 by using CT images. The combined route of viral pneumonitis feature extraction based on traditional machine vision and the COVID-19 image classification based on artificial intelligence technology has achieved a comprehensive analysis of medical image features and COVID-19 screening. The fast implementation mode of the fusion platform scenario is based on computing power and data support. Relying on the Tianhe artificial intelligence innovation-integrated service platform,the platform supports intelligent frontier innovation on the basis of computing power and data,implements an open model of simultaneous research and application,and has a multi-industry training resource model library and large-scale distributed training sources. With regard to rapid deployment and other service capabilities,this comprehensive analysis system is also the first public COVID-19 AI-assisted diagnostic system deployed online. Analysis based directly on digital imaging and communications in medicine(DICOM) data and video data will effectively improve the analysis efficiency,but it will involve data ethics and security-related issues; however,it is the developing direction that needs to be resolved in the future.关键词:Coronavirus Disease 2019(COVID-19);computed tomography(CT) imaging;auxiliary diagnosis;artificial intelligence(AI);supercomputing76|156|2更新时间:2024-05-07 -
 摘要:ObjectiveNasopharyngeal carcinoma (NPC) is a common head and neck cancer in Southeast Asia and China. In 2018, approximately 129 thousand people were diagnosed with NPC,and approximately 73 thousand people died of it. Radiotherapy has become a standard treatment method for NPC patients. Precise radiotherapy relies on the accurate delineation of tumor targets and organs-at-risk (OARs). In radiotherapy practice,these anatomical structures are usually manually delineated by radiation oncologists on a treatment-planning system (TPS). Manual delineation,however,is a time-consuming and labor-intensive process. It is also a subjective process and,hence,prone to interpractitioner variability. The NPC target segmentation is particularly challenging because of the substantial interpatient heterogeneity in tumor shape and the poorly defined tumor-to-normal tissue interface,resulting in considerable variations in gross tumor volume among physicians. Auto-segmentation methods have the potential to improve the contouring accuracy and efficiency. Different auto-segmentation methods have been reported. Nevertheless,atlas-based segmentation has long computation time and often could not account for large anatomical variations due to the uncertainty of deformable registration. Deep learning has achieved great success in computer science. It has been applied in auto-segmenting tumor targets and OARs in radiotherapy. Studies have demonstrated that the deep leaning method can perform comparably with or even better than manual segmentation for some tumor sites. In this work,we propose a Deeplabv3+ model that can automatically segment high-risk primary tumor gross target volume (GTVp) in NPC radiotherapy.MethodThe Deeplabv3+ convolutional neural network model uses an encoder-decoder structure and a spatial pyramid pooling module to complete the segmentation of high-risk primary tumor from NPC patients. The improved MobileNetV2 network is used as the network backbone,and atrous and depthwise separable convolutions are used in the encoder and decoder modules. The MobileNetV2 network consists of four inverted residual modules that contain depthwise separable convolution with striding to extract feature maps at arbitrary resolutions via atrous separable convolution. Batch normalization and ReLU activation are added after each 3×3 depthwise convolution. The decoder module of this network is as follows: the encoder features are first bilinearly upsampled by a factor of 4 and then concatenated with the corresponding low-level features from the network backbone with the same spatial resolution. We perform a 1×1 convolution on the low-level features to reduce the number of channels. After concatenation,several 3×3 convolutions are used to refine the features,followed by another bilinear upsampling by a factor of 4. Our training and test sets consist of the CT images and manual contours of 150 patients from Anhui Provincial Hospital between January 2016 and May 2019. The dimension,resolution,and thickness of CT images are 512×512,0.98 mm,and 2.5 mm,respectively. To delineate the tumor region efficiently,T1-weighted MR images are also acquired and fused with CT images. GTVp is delineated by experienced radiation oncologists on the CT images in a Pinnacle TPS. Of the 150 patients,120 are chosen as the training set,15 patients are chosen as the validation set,and the remaining 15 patients are chosen as the test set. Images are flipped,translated,and randomly rotated to augment the training dataset. Our network is implemented in Keras toolbox. The input images and ground-truth contours are resized to 512×512 for training. The loss function used in this study is 1-DSC index,AdamOptimizer is used with a learning rate of 0.005,and the weight decay factor is 0.8. The performance of the auto-segmentation algorithm is evaluated with Dice similarity coefficient (DSC),Jaccard index (JI),average surface distance (ASD),and Hausdorff distance (HD). The results are compared with those of the U-Net model. Paired t-test is performed to compare the DSC,JI,ASD,and HD values between the different models.ResultThe mean DSC value of the 15 NPC patients from the test set is 0.76±0.11,the mean JI value is 0.63±0.13,the average ASD value is (3.4±2.0) mm,and the average HD value is (10.9±8.6) mm. Compared with the U-Net model,the Deeplabv3+ network model shows improved mean DSC and JI values by 3% and 4%,respectively (0.76±0.11 vs. 0.73±0.13,p < 0.001; 0.63±0.13 vs. 0.59±0.14,p < 0.001). The mean ASD value is also significantly reduced (3.4±2.0 vs. 3.8±3.3 mm,p=0.014) compared with the U-Net result. However,for HD values,no statistical difference exists between the two network models (10.9±8.6 vs. 11.1±7.5 mm,p=0.745). The experiment results indicate that the Deeplabv3+ network model outperforms the U-Net model in the segmentation of NPC target area. As 2D visualizations of auto-segmented contours,the Deeplabv3+ model results have more overlap with the manual contours and are closer to the results of the "ground truth". The visualizations show that our model can produce refined results. In addition,the average time required to segment a CT image is 16 and 14 ms for our model and the U-Net model,respectively,which is much less than the manual contouring time.ConclusionIn this study,a Deeplabv3+ convolutional neural network model is proposed to auto-segment the GTVp of NPC patients with radiotherapy. The results show that the auto-segmentations of the Deeplabv3+ network are close to the manual contours from oncologists. This model has the potential to improve the efficiency and consistency of GTVp contouring for NPC patients.关键词:auto-segmentation;radiotherapy;convolutional neural network(CNN);primary tumor gross target volume (GTVp);nasopharyngeal carcinoma(NPC)31|27|0更新时间:2024-05-07
摘要:ObjectiveNasopharyngeal carcinoma (NPC) is a common head and neck cancer in Southeast Asia and China. In 2018, approximately 129 thousand people were diagnosed with NPC,and approximately 73 thousand people died of it. Radiotherapy has become a standard treatment method for NPC patients. Precise radiotherapy relies on the accurate delineation of tumor targets and organs-at-risk (OARs). In radiotherapy practice,these anatomical structures are usually manually delineated by radiation oncologists on a treatment-planning system (TPS). Manual delineation,however,is a time-consuming and labor-intensive process. It is also a subjective process and,hence,prone to interpractitioner variability. The NPC target segmentation is particularly challenging because of the substantial interpatient heterogeneity in tumor shape and the poorly defined tumor-to-normal tissue interface,resulting in considerable variations in gross tumor volume among physicians. Auto-segmentation methods have the potential to improve the contouring accuracy and efficiency. Different auto-segmentation methods have been reported. Nevertheless,atlas-based segmentation has long computation time and often could not account for large anatomical variations due to the uncertainty of deformable registration. Deep learning has achieved great success in computer science. It has been applied in auto-segmenting tumor targets and OARs in radiotherapy. Studies have demonstrated that the deep leaning method can perform comparably with or even better than manual segmentation for some tumor sites. In this work,we propose a Deeplabv3+ model that can automatically segment high-risk primary tumor gross target volume (GTVp) in NPC radiotherapy.MethodThe Deeplabv3+ convolutional neural network model uses an encoder-decoder structure and a spatial pyramid pooling module to complete the segmentation of high-risk primary tumor from NPC patients. The improved MobileNetV2 network is used as the network backbone,and atrous and depthwise separable convolutions are used in the encoder and decoder modules. The MobileNetV2 network consists of four inverted residual modules that contain depthwise separable convolution with striding to extract feature maps at arbitrary resolutions via atrous separable convolution. Batch normalization and ReLU activation are added after each 3×3 depthwise convolution. The decoder module of this network is as follows: the encoder features are first bilinearly upsampled by a factor of 4 and then concatenated with the corresponding low-level features from the network backbone with the same spatial resolution. We perform a 1×1 convolution on the low-level features to reduce the number of channels. After concatenation,several 3×3 convolutions are used to refine the features,followed by another bilinear upsampling by a factor of 4. Our training and test sets consist of the CT images and manual contours of 150 patients from Anhui Provincial Hospital between January 2016 and May 2019. The dimension,resolution,and thickness of CT images are 512×512,0.98 mm,and 2.5 mm,respectively. To delineate the tumor region efficiently,T1-weighted MR images are also acquired and fused with CT images. GTVp is delineated by experienced radiation oncologists on the CT images in a Pinnacle TPS. Of the 150 patients,120 are chosen as the training set,15 patients are chosen as the validation set,and the remaining 15 patients are chosen as the test set. Images are flipped,translated,and randomly rotated to augment the training dataset. Our network is implemented in Keras toolbox. The input images and ground-truth contours are resized to 512×512 for training. The loss function used in this study is 1-DSC index,AdamOptimizer is used with a learning rate of 0.005,and the weight decay factor is 0.8. The performance of the auto-segmentation algorithm is evaluated with Dice similarity coefficient (DSC),Jaccard index (JI),average surface distance (ASD),and Hausdorff distance (HD). The results are compared with those of the U-Net model. Paired t-test is performed to compare the DSC,JI,ASD,and HD values between the different models.ResultThe mean DSC value of the 15 NPC patients from the test set is 0.76±0.11,the mean JI value is 0.63±0.13,the average ASD value is (3.4±2.0) mm,and the average HD value is (10.9±8.6) mm. Compared with the U-Net model,the Deeplabv3+ network model shows improved mean DSC and JI values by 3% and 4%,respectively (0.76±0.11 vs. 0.73±0.13,p < 0.001; 0.63±0.13 vs. 0.59±0.14,p < 0.001). The mean ASD value is also significantly reduced (3.4±2.0 vs. 3.8±3.3 mm,p=0.014) compared with the U-Net result. However,for HD values,no statistical difference exists between the two network models (10.9±8.6 vs. 11.1±7.5 mm,p=0.745). The experiment results indicate that the Deeplabv3+ network model outperforms the U-Net model in the segmentation of NPC target area. As 2D visualizations of auto-segmented contours,the Deeplabv3+ model results have more overlap with the manual contours and are closer to the results of the "ground truth". The visualizations show that our model can produce refined results. In addition,the average time required to segment a CT image is 16 and 14 ms for our model and the U-Net model,respectively,which is much less than the manual contouring time.ConclusionIn this study,a Deeplabv3+ convolutional neural network model is proposed to auto-segment the GTVp of NPC patients with radiotherapy. The results show that the auto-segmentations of the Deeplabv3+ network are close to the manual contours from oncologists. This model has the potential to improve the efficiency and consistency of GTVp contouring for NPC patients.关键词:auto-segmentation;radiotherapy;convolutional neural network(CNN);primary tumor gross target volume (GTVp);nasopharyngeal carcinoma(NPC)31|27|0更新时间:2024-05-07 -
 摘要:ObjectiveArteriovenous (A/V) classification in fundus images is a fundamental step for the risk assessment of many systemic diseases. A/V classification methods based on traditional machine learning require complicated feature engineering, consistently rely on the results of blood vessel extraction, and cannot achieve end-to-end A/V classification. The development of deep semantic segmentation technology makes end-to-end A/V classification possible, and has been commonly used in fundus image analysis. In this paper, a segmentation model semantic fusion based U-Net (SFU-Net) is proposed combined with the powerful feature extraction capabilities of deep learning to improve the accuracy of A/V classification.MethodFirst, the arteries and veins in the fundus image belong to blood vessels and are highly similar in structure. Existing deep learning-based A/V classification methods frequently treat this problem as a multiclassification problem. This paper proposes a multilabel learning strategy to address this problem for reducing the difficulty of optimization and deal with the situation where the arteries and veins in the fundus image cross. The lower layers of the network are mainly responsible for extracting the common features of the two structures. The upper layers of the network learn two binary classifiers and extract the arteries and veins independently. Second, considering the high similarity of the description features of arteries and veins in color and structure, this paper improves the U-Net architecture in two aspects. 1) The original simple feature extractor of U-Net is replaced by DenseNet-121. The original U-Net encoder is composed of 10 convolutional layers and four maximum pooling layers, and the feature extraction capability is extremely limited. By contrast, DenseNet-121 has many convolutional layers, and the introduction of dense connections makes the feature utilization rate high, the transmission efficiency of features and gradients in the network is high, and the feature extraction ability is strong. This paper reduces four downsampling operations of U-Net to three, and the input image is downsampled by eight times to avoid the loss of detailed information. 2) A semantic fusion module is proposed. The semantic fusion module includes two operations, namely, feature fusion and channelwise attention mechanism. Low-level features have high resolution and contain many location and detail information, but few semantic features and many noises. High-level features have strong semantic information, but their resolution is extremely low and the detail information is few. The features from different layers are first fused to enhance their distinguishing ability. For the fused features, the channelwise attention mechanism is used to select the features. The convolution filter can only capture local information. The global average pooling operation is performed on the input features in the channel dimension to capture global context information. Each element of the resulting vector is a concentrated representation of its corresponding channel. Two nonlinear transformations are then performed on the vector to model the correlation between channels and reduce the amount of parameters and calculations. The vector is restored to its original dimension and normalized to 0-1 through the sigmoid gate. Each element in the obtained vector is regarded as the importance of each channel in the input feature, and each feature channel of the input feature is weighted through a multiplication operation. Through the channel attention mechanism, the network can automatically focus on the feature channels that are important to the task while suppressing the features that are unimportant, thereby improving the performance of the model during the training process. Third, considering the problem of uneven distribution between blood vessels and background pixels in the fundus image, this paper takes focal loss as the loss function to solve the problem of class imbalance and focus on difficult samples at the same time. Focal loss introduces parameters α and γ in the cross-entropy loss function. Parameter α is used to balance the difference between positive and negative samples. Parameter γ adjusts the degree where the loss of simple samples is reduced, thereby amplifying the difference between the loss values of difficult and simple samples. The values of the two parameters are determined through cross-validation. The overall optimization goal is the sum of the focal loss of arteries and veins, thereby optimizing the arteries and veins during training.ResultThe proposed method is verified on two public datasets, namely, digital retinal images for vessel extraction(DRIVE) and WIDE, and its performance is evaluated from two perspectives, namely, segmentation and classification. Experimental results demonstrate that the proposed method shows better performance than most existing methods. The proposed method achieves an area under the curve of 0.968 6 and 0.973 6 for segmenting arteries and veins on the DRIVE dataset, and the sensitivity, specificity, accuracy and balanced-accuracy of A/V classification are 88.39%, 94.25%, 91.68%, and 91.32%, respectively. Compared with state-of-the-art method, the sensitivity of the proposed method only decreases by 0.61%, and specificity, accuracy, and balaned-auuracy have absolute improvements of 4.25%, 2.68%, and 1.82%, respectively. The proposed method achieves an accuracy of 92.38%, which is 6.18% higher than the state-of-the-art method.ConclusionThe fusion module can effectively use multi-scale features and automatically select many important features, thereby improving performance. The proposed method performs well in A/V classification, exceeding most existing methods.关键词:fundus images;arteriovenous classification;deep learning;semantic segmentation;feature fusion50|84|2更新时间:2024-05-07
摘要:ObjectiveArteriovenous (A/V) classification in fundus images is a fundamental step for the risk assessment of many systemic diseases. A/V classification methods based on traditional machine learning require complicated feature engineering, consistently rely on the results of blood vessel extraction, and cannot achieve end-to-end A/V classification. The development of deep semantic segmentation technology makes end-to-end A/V classification possible, and has been commonly used in fundus image analysis. In this paper, a segmentation model semantic fusion based U-Net (SFU-Net) is proposed combined with the powerful feature extraction capabilities of deep learning to improve the accuracy of A/V classification.MethodFirst, the arteries and veins in the fundus image belong to blood vessels and are highly similar in structure. Existing deep learning-based A/V classification methods frequently treat this problem as a multiclassification problem. This paper proposes a multilabel learning strategy to address this problem for reducing the difficulty of optimization and deal with the situation where the arteries and veins in the fundus image cross. The lower layers of the network are mainly responsible for extracting the common features of the two structures. The upper layers of the network learn two binary classifiers and extract the arteries and veins independently. Second, considering the high similarity of the description features of arteries and veins in color and structure, this paper improves the U-Net architecture in two aspects. 1) The original simple feature extractor of U-Net is replaced by DenseNet-121. The original U-Net encoder is composed of 10 convolutional layers and four maximum pooling layers, and the feature extraction capability is extremely limited. By contrast, DenseNet-121 has many convolutional layers, and the introduction of dense connections makes the feature utilization rate high, the transmission efficiency of features and gradients in the network is high, and the feature extraction ability is strong. This paper reduces four downsampling operations of U-Net to three, and the input image is downsampled by eight times to avoid the loss of detailed information. 2) A semantic fusion module is proposed. The semantic fusion module includes two operations, namely, feature fusion and channelwise attention mechanism. Low-level features have high resolution and contain many location and detail information, but few semantic features and many noises. High-level features have strong semantic information, but their resolution is extremely low and the detail information is few. The features from different layers are first fused to enhance their distinguishing ability. For the fused features, the channelwise attention mechanism is used to select the features. The convolution filter can only capture local information. The global average pooling operation is performed on the input features in the channel dimension to capture global context information. Each element of the resulting vector is a concentrated representation of its corresponding channel. Two nonlinear transformations are then performed on the vector to model the correlation between channels and reduce the amount of parameters and calculations. The vector is restored to its original dimension and normalized to 0-1 through the sigmoid gate. Each element in the obtained vector is regarded as the importance of each channel in the input feature, and each feature channel of the input feature is weighted through a multiplication operation. Through the channel attention mechanism, the network can automatically focus on the feature channels that are important to the task while suppressing the features that are unimportant, thereby improving the performance of the model during the training process. Third, considering the problem of uneven distribution between blood vessels and background pixels in the fundus image, this paper takes focal loss as the loss function to solve the problem of class imbalance and focus on difficult samples at the same time. Focal loss introduces parameters α and γ in the cross-entropy loss function. Parameter α is used to balance the difference between positive and negative samples. Parameter γ adjusts the degree where the loss of simple samples is reduced, thereby amplifying the difference between the loss values of difficult and simple samples. The values of the two parameters are determined through cross-validation. The overall optimization goal is the sum of the focal loss of arteries and veins, thereby optimizing the arteries and veins during training.ResultThe proposed method is verified on two public datasets, namely, digital retinal images for vessel extraction(DRIVE) and WIDE, and its performance is evaluated from two perspectives, namely, segmentation and classification. Experimental results demonstrate that the proposed method shows better performance than most existing methods. The proposed method achieves an area under the curve of 0.968 6 and 0.973 6 for segmenting arteries and veins on the DRIVE dataset, and the sensitivity, specificity, accuracy and balanced-accuracy of A/V classification are 88.39%, 94.25%, 91.68%, and 91.32%, respectively. Compared with state-of-the-art method, the sensitivity of the proposed method only decreases by 0.61%, and specificity, accuracy, and balaned-auuracy have absolute improvements of 4.25%, 2.68%, and 1.82%, respectively. The proposed method achieves an accuracy of 92.38%, which is 6.18% higher than the state-of-the-art method.ConclusionThe fusion module can effectively use multi-scale features and automatically select many important features, thereby improving performance. The proposed method performs well in A/V classification, exceeding most existing methods.关键词:fundus images;arteriovenous classification;deep learning;semantic segmentation;feature fusion50|84|2更新时间:2024-05-07 -
 摘要:ObjectiveTwenty-three pairs of chromosomes, including 22 pairs of autosomes and a pair of sex chromosomes, are found in the cells of a healthy human body. As those chromosomes are vital genetic information carriers, karyotyping chromosomes is important for medical diagnostics, drug development, and biomedical research. Chromosome karyotyping analysis refers to segmenting chromosome instances from a stained cell microphotograph and arranging them into karyotype in accordance with their band criterion. However, chromosome karyotyping analysis is usually performed by skilled cytologists manually, which requires extensive experience, domain expertise, and considerable manual efforts. In this study, we focus on the segmentation of chromosome instances because it is a crucial and challenging obstacle of chromosome karyotyping.MethodIn this study, we propose the AS-PANet(amount segmentation PANet) method by improving the PANet instance segmentation model for the segmentation task of chromosome instances. We add a chromosome counting task into AS-PANet for joint training, thereby enabling the classification, detection, segmentation, and counting tasks to share latent features. We propose a chromosome label augmentation algorithm for augmenting the training dataset.ResultWe collect clinical metaphase cell microphotograph images from Guangdong Women and Children Hospital. Then, we build a clinical dataset by labeling 802 chromosome clusters. The clinical dataset is split into training and testing datasets with a ratio of 8 :2. The proposed augmentation algorithm augments the training dataset from 802 samples into 13 482 samples. Our solution achieves a more competitive result on the clinical chromosome dataset with 90.63% mean average precision (mAP), whereas PANet yields 89.45% mAP and Mask R-CNN(regirn with convolutional neural network) obtains 87.78% mAP at the equal experimental condition. The AS-PANet method yields 85% instance segmentation accuracy, which is 2% higher than the result of PANet and 3.75% higher than the result of Mask R-CNN.ConclusionExperimental results demonstrate that the proposed method is an effective and promising solution for solving the segmentation problem of clinical chromosome instances. The contributions and highlights of this study are summarized as follows: 1) we proposed a novel chromosome instance segmentation method by improving the path aggregation network architecture. 2) We built a clinical dataset for training and verifying the proposed method. 3) We demonstrated the effectiveness of the proposed method for tackling the chromosome instance segmentation task. Experimental results showed that the proposed method is more promising than previous studies. 4) The highlights of this work are high chromosome segmentation certainty and accuracy with a small amount of manual labeling cost.关键词:amount segmentation PANet(AS-PANet);path aggregation network(PANet);chromosome segmentation;instance segmentation;chromosome karyotyping analysis92|112|9更新时间:2024-05-07
摘要:ObjectiveTwenty-three pairs of chromosomes, including 22 pairs of autosomes and a pair of sex chromosomes, are found in the cells of a healthy human body. As those chromosomes are vital genetic information carriers, karyotyping chromosomes is important for medical diagnostics, drug development, and biomedical research. Chromosome karyotyping analysis refers to segmenting chromosome instances from a stained cell microphotograph and arranging them into karyotype in accordance with their band criterion. However, chromosome karyotyping analysis is usually performed by skilled cytologists manually, which requires extensive experience, domain expertise, and considerable manual efforts. In this study, we focus on the segmentation of chromosome instances because it is a crucial and challenging obstacle of chromosome karyotyping.MethodIn this study, we propose the AS-PANet(amount segmentation PANet) method by improving the PANet instance segmentation model for the segmentation task of chromosome instances. We add a chromosome counting task into AS-PANet for joint training, thereby enabling the classification, detection, segmentation, and counting tasks to share latent features. We propose a chromosome label augmentation algorithm for augmenting the training dataset.ResultWe collect clinical metaphase cell microphotograph images from Guangdong Women and Children Hospital. Then, we build a clinical dataset by labeling 802 chromosome clusters. The clinical dataset is split into training and testing datasets with a ratio of 8 :2. The proposed augmentation algorithm augments the training dataset from 802 samples into 13 482 samples. Our solution achieves a more competitive result on the clinical chromosome dataset with 90.63% mean average precision (mAP), whereas PANet yields 89.45% mAP and Mask R-CNN(regirn with convolutional neural network) obtains 87.78% mAP at the equal experimental condition. The AS-PANet method yields 85% instance segmentation accuracy, which is 2% higher than the result of PANet and 3.75% higher than the result of Mask R-CNN.ConclusionExperimental results demonstrate that the proposed method is an effective and promising solution for solving the segmentation problem of clinical chromosome instances. The contributions and highlights of this study are summarized as follows: 1) we proposed a novel chromosome instance segmentation method by improving the path aggregation network architecture. 2) We built a clinical dataset for training and verifying the proposed method. 3) We demonstrated the effectiveness of the proposed method for tackling the chromosome instance segmentation task. Experimental results showed that the proposed method is more promising than previous studies. 4) The highlights of this work are high chromosome segmentation certainty and accuracy with a small amount of manual labeling cost.关键词:amount segmentation PANet(AS-PANet);path aggregation network(PANet);chromosome segmentation;instance segmentation;chromosome karyotyping analysis92|112|9更新时间:2024-05-07
Computerd Tomography Image
-
 摘要:ObjectiveBrain tumor is a serious threat to human health. The invasive growth of brain tumor,when it occupies a certain space in the skull,will lead to increased intracranial pressure and compression of brain tissue,which will damage the central nerve and even threaten the patient's life. Therefore,effective brain tumor diagnosis and timely treatment are of great significance to improving the patient's quality of life and prolonging the patient's life. Computer-assisted segmentation of brain tumor is necessary for the prognosis and treatment of patients. However,although brain-related research has made great progress,automatic identification of the contour information of tumor and effective segmentation of each subregion in MRI(magnetic resonance imaging) remain difficult due to the highly heterogeneous appearance,random location,and large difference in the number of voxels in each subregion of the tumor and the high degree of gray-scale similarity between the tumor tissue and neighboring normal brain tissue. Since 2012, with the development of deep learning and the improvement of related hardware performance,segmentation methods based on neural networks have gradually become the mainstream. In particular,3D convolutional neural networks are widely used in the field of brain tumor segmentation because of their advantages of sufficient spatial feature extraction and high segmentation effect. Nonetheless,their large memory consumption and high requirements on hardware resources usually require making a compromise in the network structure that adapts to the given memory budget at the expense of accuracy or training speed. To address such problems,we propose a lightweight segmentation algorithm in this paper.MethodFirst,group convolution was used to replace conventional convolution for significantly reducing the parameters and improving segmentation accuracy because memory consumption is negatively correlated with batch size. A large batch size usually means enhanced convergence stability and training effect in 3D convolutional neural networks. Then,multifiber and channel shuffle units were used to enhance the information fusion among the groups and compensate for the poor communication caused by group convolution. Synchronized cross-GPU batch normalization was used to alleviate the poor training performance of 3D convolutional neural networks due to the small batch size and utilize the advantages of multigraphics collaborative computing. Aiming at the case in which the subregions have different difficulties in segmentation,a weighted mixed-loss function consisting of Dice and Jaccard losses was proposed to improve the segmentation accuracy of the subregions that are difficult to segment under the premise of maintaining the high precision of the easily segmented subregions and accelerate the model convergence speed. One of the most challenging parts of the task is to distinguish between small blood vessels in the tumor core and enhanced-tumor areas. This process is particularly difficult for the labels that may not have enhanced tumor at all. If neither the ground truth nor the prediction has an enhanced area,the Dice score of the enhancement area is 1. Conversely,in patients who did not have enhanced tumors in the ground truth,only a single false-positive voxel would result in a Dice score of 0. Hence,we postprocessed the prediction results,that is,we set a threshold for the number of voxels in the tumor-enhanced area. When the number of voxels in the tumor-enhanced area is less than the threshold,these voxels would be merged into the tumor core area,thereby improving the Dice score of the tumor-enhanced and tumor core areas.ResultTo verify the overall performance of the algorithm,we first conducted a fivefold cross-validation evaluation on the training set of the public brain tumor dataset BraTS2018. The average Dice scores of the proposed algorithm in the entire tumor,tumor core,and enhanced tumor areas can reach 89.52%,82.74%,and 77.19%,respectively. For fairness,an experiment was also conducted on the BraTS2018 validation set. We used the trained network to segment the unlabeled samples for prediction,converted them into the corresponding format,and uploaded them to the BraTS online server. The segmentation results were provided by the server after calculation and analysis. The proposed algorithm achieves average Dice scores of 90.67%,85.06%,and 80.41%. The parameters and floating point operations are 3.2 M and 20.51 G,respectively. Compared with the classic 3D U-Net,our algorithm shows higher average Dice scores by 2.14%,13.29%,and 4.45%. Moreover,the parameters and floating point operations are reduced by 5 and 81 times,respectively. Compared with the state-of-the-art approach that won the first place in the 2018 Multimodal Brain Tumor Segmentation Challenge,the average Dice scores are reduced by only 0.01%,0.96%,and 1.32%. Nevertheless,the parameters and floating point operations are reduced by 12 and 73 times,respectively,indicating a more practical value.ConclusionAiming at the problems of large memory consumption and slow segmentation speed in the field of computer-aided brain tumor segmentation,an algorithm combining group convolution and channel shuffle unit is proposed. The punishment intensity of sparse class classification error to model is improved using the weighted mixed loss function to balance the training intensity of different segmentation difficulty categories effectively. The experimental results show that the algorithm can significantly reduce the computational cost while maintaining high accuracy and provide a powerful reference for clinicians in brain tumor segmentation.关键词:magnetic resonance imaging(MRI);brain tumor segmentation;deep learning;group convolution;weighted mixed loss function51|74|2更新时间:2024-05-07
摘要:ObjectiveBrain tumor is a serious threat to human health. The invasive growth of brain tumor,when it occupies a certain space in the skull,will lead to increased intracranial pressure and compression of brain tissue,which will damage the central nerve and even threaten the patient's life. Therefore,effective brain tumor diagnosis and timely treatment are of great significance to improving the patient's quality of life and prolonging the patient's life. Computer-assisted segmentation of brain tumor is necessary for the prognosis and treatment of patients. However,although brain-related research has made great progress,automatic identification of the contour information of tumor and effective segmentation of each subregion in MRI(magnetic resonance imaging) remain difficult due to the highly heterogeneous appearance,random location,and large difference in the number of voxels in each subregion of the tumor and the high degree of gray-scale similarity between the tumor tissue and neighboring normal brain tissue. Since 2012, with the development of deep learning and the improvement of related hardware performance,segmentation methods based on neural networks have gradually become the mainstream. In particular,3D convolutional neural networks are widely used in the field of brain tumor segmentation because of their advantages of sufficient spatial feature extraction and high segmentation effect. Nonetheless,their large memory consumption and high requirements on hardware resources usually require making a compromise in the network structure that adapts to the given memory budget at the expense of accuracy or training speed. To address such problems,we propose a lightweight segmentation algorithm in this paper.MethodFirst,group convolution was used to replace conventional convolution for significantly reducing the parameters and improving segmentation accuracy because memory consumption is negatively correlated with batch size. A large batch size usually means enhanced convergence stability and training effect in 3D convolutional neural networks. Then,multifiber and channel shuffle units were used to enhance the information fusion among the groups and compensate for the poor communication caused by group convolution. Synchronized cross-GPU batch normalization was used to alleviate the poor training performance of 3D convolutional neural networks due to the small batch size and utilize the advantages of multigraphics collaborative computing. Aiming at the case in which the subregions have different difficulties in segmentation,a weighted mixed-loss function consisting of Dice and Jaccard losses was proposed to improve the segmentation accuracy of the subregions that are difficult to segment under the premise of maintaining the high precision of the easily segmented subregions and accelerate the model convergence speed. One of the most challenging parts of the task is to distinguish between small blood vessels in the tumor core and enhanced-tumor areas. This process is particularly difficult for the labels that may not have enhanced tumor at all. If neither the ground truth nor the prediction has an enhanced area,the Dice score of the enhancement area is 1. Conversely,in patients who did not have enhanced tumors in the ground truth,only a single false-positive voxel would result in a Dice score of 0. Hence,we postprocessed the prediction results,that is,we set a threshold for the number of voxels in the tumor-enhanced area. When the number of voxels in the tumor-enhanced area is less than the threshold,these voxels would be merged into the tumor core area,thereby improving the Dice score of the tumor-enhanced and tumor core areas.ResultTo verify the overall performance of the algorithm,we first conducted a fivefold cross-validation evaluation on the training set of the public brain tumor dataset BraTS2018. The average Dice scores of the proposed algorithm in the entire tumor,tumor core,and enhanced tumor areas can reach 89.52%,82.74%,and 77.19%,respectively. For fairness,an experiment was also conducted on the BraTS2018 validation set. We used the trained network to segment the unlabeled samples for prediction,converted them into the corresponding format,and uploaded them to the BraTS online server. The segmentation results were provided by the server after calculation and analysis. The proposed algorithm achieves average Dice scores of 90.67%,85.06%,and 80.41%. The parameters and floating point operations are 3.2 M and 20.51 G,respectively. Compared with the classic 3D U-Net,our algorithm shows higher average Dice scores by 2.14%,13.29%,and 4.45%. Moreover,the parameters and floating point operations are reduced by 5 and 81 times,respectively. Compared with the state-of-the-art approach that won the first place in the 2018 Multimodal Brain Tumor Segmentation Challenge,the average Dice scores are reduced by only 0.01%,0.96%,and 1.32%. Nevertheless,the parameters and floating point operations are reduced by 12 and 73 times,respectively,indicating a more practical value.ConclusionAiming at the problems of large memory consumption and slow segmentation speed in the field of computer-aided brain tumor segmentation,an algorithm combining group convolution and channel shuffle unit is proposed. The punishment intensity of sparse class classification error to model is improved using the weighted mixed loss function to balance the training intensity of different segmentation difficulty categories effectively. The experimental results show that the algorithm can significantly reduce the computational cost while maintaining high accuracy and provide a powerful reference for clinicians in brain tumor segmentation.关键词:magnetic resonance imaging(MRI);brain tumor segmentation;deep learning;group convolution;weighted mixed loss function51|74|2更新时间:2024-05-07 -
 摘要:ObjectiveMagnetic resonance imaging (MRI) is frequently used in clinical applications. It is a common means to detect lesions,injuries,and soft tissue variations in neural system diseases. Skull removal is an important preprocessing step for brain magnetic resonance (MR) image analysis. Its purpose is to remove nonbrain tissue from the brain MRI,thereby facilitating subsequent extraction and analysis of brain tissue. The MR images acquired using clinical scanners inevitably have blurring or noise characteristics due to the complexity of brain tissue structure and the effects of equipment noise and field offset. Differences also exist in the anatomical structure of the brain tissue for different individuals,which cause difficulties in the skull segmentation in brain MR images. Most traditional methods for skull segmentation are incompletely automatic and often require the operator to use the mouse and other tools to determine the center point of the region of interest and adjust the parameters manually. The current automatic skull segmentation method does not require human-computer interaction but has poor adaptability,and satisfactory segmentation results in different MR images are difficult to achieve. On the contrary,the deep learning-based method exhibits advanced performance in multiple segmentation tasks in the field of computer vision. Therefore,we propose a deep iterative fusion convolutional neural network model (DIFNet) in this work to realize skull segmentation.MethodThe main structure of DIFNet is composed of an encoder and a decoder. The skip connection between the encoder and decoder is realized by multiple upsampling iterative fusion,which means that the input information of one decoder layer comes from not only the same layer but also the deep layers of the encoder. The encoder consists of several residual convolution blocks,which allow the shallow semantic information to flow into deep networks to avoid gradient vanishment. The decoder is composed of double-way upsampling modules. The feature maps generated from the double-way upsampling modules are added as real outputs through deconvolution operations with different receptive field sizes. This process enables to restore the image details effectively by adding multiple scale information. The internal data enhancement method is adopted to enhance the generalization capability of the model. First,the image is randomly scaled,in which the interval of scaling factor sets is determined in accordance with the ratio of the original image size to the output block size. Then,a center point is randomly selected in the scaled image,and the cutting area is determined. Lastly,the cut image patches are fed into the network for training. The Dice loss function embedded with an L2 regularization item is used to optimize the model parameters and overcome the overfitting problem. We use two datasets in this work to evaluate the accuracy and robustness of the proposed model. Each dataset has a brain segmentation mask provided by a professional doctor as the gold standard of the model. One dataset is NFBS(neurofeed back skullstripped),from which a part of images are used for testing (the ratio of the training dataset to the test dataset is 4 :1). The other dataset is LPBA40(loni probabilistic brain atlas 40),which is used as an independent dataset for testing the generality of the models. For quantitative analysis,the Dice score,sensitivity,and specificity are used in this work.ResultFor the NFBS dataset,the method in this paper obtains the highest average Dice score and sensitivity of 99.12% and 99.22%,respectively,compared with U-Net,U-Net with residual block (Res-U-Net),and U-Net with double-way upsampling modules (UP-U-Net). The Dice score is increased by 1.88%,1.81%,and 0.6%. The sensitivity and septicity are increased by at least 0.5% compared with the U-Net model. The segmentation results of the model are similar to the manual segmentation results of experts. The model trained with the NFBS dataset is applied directly to the LPBA40 dataset to verify the segmentation capability of the model. The Dice value obtained in the test experiment is up to 98.16%. By contrast,the Dice values of U-Net,UP-U-Net,and Res-U-Net are 81.69%,77.34%,and 76.42%,respectively. Compared with these models,our proposed model is robust.ConclusionExperiments illustrate that the internal data augmentation and deep iterative fusion make the proposed model be easily trained and acquire the best segmentation results. The deep iterative feature fusion can guarantee the robustness of the segmentation model.关键词:convolutional neural network(CNN);skull segmentation;brain magnetic resonance image;deep iterative fusion;data augmentation71|189|0更新时间:2024-05-07
摘要:ObjectiveMagnetic resonance imaging (MRI) is frequently used in clinical applications. It is a common means to detect lesions,injuries,and soft tissue variations in neural system diseases. Skull removal is an important preprocessing step for brain magnetic resonance (MR) image analysis. Its purpose is to remove nonbrain tissue from the brain MRI,thereby facilitating subsequent extraction and analysis of brain tissue. The MR images acquired using clinical scanners inevitably have blurring or noise characteristics due to the complexity of brain tissue structure and the effects of equipment noise and field offset. Differences also exist in the anatomical structure of the brain tissue for different individuals,which cause difficulties in the skull segmentation in brain MR images. Most traditional methods for skull segmentation are incompletely automatic and often require the operator to use the mouse and other tools to determine the center point of the region of interest and adjust the parameters manually. The current automatic skull segmentation method does not require human-computer interaction but has poor adaptability,and satisfactory segmentation results in different MR images are difficult to achieve. On the contrary,the deep learning-based method exhibits advanced performance in multiple segmentation tasks in the field of computer vision. Therefore,we propose a deep iterative fusion convolutional neural network model (DIFNet) in this work to realize skull segmentation.MethodThe main structure of DIFNet is composed of an encoder and a decoder. The skip connection between the encoder and decoder is realized by multiple upsampling iterative fusion,which means that the input information of one decoder layer comes from not only the same layer but also the deep layers of the encoder. The encoder consists of several residual convolution blocks,which allow the shallow semantic information to flow into deep networks to avoid gradient vanishment. The decoder is composed of double-way upsampling modules. The feature maps generated from the double-way upsampling modules are added as real outputs through deconvolution operations with different receptive field sizes. This process enables to restore the image details effectively by adding multiple scale information. The internal data enhancement method is adopted to enhance the generalization capability of the model. First,the image is randomly scaled,in which the interval of scaling factor sets is determined in accordance with the ratio of the original image size to the output block size. Then,a center point is randomly selected in the scaled image,and the cutting area is determined. Lastly,the cut image patches are fed into the network for training. The Dice loss function embedded with an L2 regularization item is used to optimize the model parameters and overcome the overfitting problem. We use two datasets in this work to evaluate the accuracy and robustness of the proposed model. Each dataset has a brain segmentation mask provided by a professional doctor as the gold standard of the model. One dataset is NFBS(neurofeed back skullstripped),from which a part of images are used for testing (the ratio of the training dataset to the test dataset is 4 :1). The other dataset is LPBA40(loni probabilistic brain atlas 40),which is used as an independent dataset for testing the generality of the models. For quantitative analysis,the Dice score,sensitivity,and specificity are used in this work.ResultFor the NFBS dataset,the method in this paper obtains the highest average Dice score and sensitivity of 99.12% and 99.22%,respectively,compared with U-Net,U-Net with residual block (Res-U-Net),and U-Net with double-way upsampling modules (UP-U-Net). The Dice score is increased by 1.88%,1.81%,and 0.6%. The sensitivity and septicity are increased by at least 0.5% compared with the U-Net model. The segmentation results of the model are similar to the manual segmentation results of experts. The model trained with the NFBS dataset is applied directly to the LPBA40 dataset to verify the segmentation capability of the model. The Dice value obtained in the test experiment is up to 98.16%. By contrast,the Dice values of U-Net,UP-U-Net,and Res-U-Net are 81.69%,77.34%,and 76.42%,respectively. Compared with these models,our proposed model is robust.ConclusionExperiments illustrate that the internal data augmentation and deep iterative fusion make the proposed model be easily trained and acquire the best segmentation results. The deep iterative feature fusion can guarantee the robustness of the segmentation model.关键词:convolutional neural network(CNN);skull segmentation;brain magnetic resonance image;deep iterative fusion;data augmentation71|189|0更新时间:2024-05-07 -
 摘要:ObjectiveMedical image registration is an important research task in the field of medical image analysis,which is the basis of medical image fusion and medical image reconstruction. Conventional registration methods that build loss function based on normalized mutual information by using iterative gradient descent to achieve registration tend to be time consuming. The existing deep learning-based medical image registration methods have limitation in registering medical images with large non-rigid deformation,which cannot achieve high registration accuracy and have poor generalization ability. Thus,this paper proposes a method to register multi-modal medical images by combining residual-in-residual dense block (RRDB) with generative adversarial network (GAN).MethodFirst,RRDB are introduced to the standard generator network to extract high-level feature information from unpaired image pairs; thus,registration accuracy is improved. Then,a least-squares loss is used to substitute cross-entropy loss constructed by the logistic regression objective. The convergence condition of least-squares loss is strict and promotes the model to reach convergence at the optimal parameters,which can alleviate gradient disappearance and overfitting; thus,the robustness of model training is enhanced. In addition,relative average GAN (RaGAN) is embedded into the standard discriminator network,namely,adding a gradient penalty in the discriminator network,which reduce the error of discriminator to estimate the relative true and false probability between the fixed image and moving image. The enhanced discriminator can help the generator to learn clearer and texture information; therefore,the registration error can be decreased,and the registration accuracy can be stabilized.ResultThis registration model is trained and validated on DRIVE(digital retinal images for vessel extraction) dataset. Generalization performance tests are performed on Sunybrook Cardiac Dataset and Brain MRI Dataset. Compared with state-of-the-art registration methods,extensive experiments demonstrate that the proposed model achieves good registration results; both registration accuracy and generalization ability have been improved. Compared with the basic literature,the registration Dice values of retinal images,cardiac images,and brain images are improved by 3.3%,3.0%,and 1.5%,respectively. According to the stability verification experiment of the registration model,as the number of iterations augments,the Dice value of this paper gradually increases,and the change is more stable than that of baseline literature. The number of iterations in this paper is 80 000,whereas that of baseline literature is 10 000. This verification experiment shows that this registration model has been effectively improved in the training phase; not only the convergence speed is accelerated but also stability is higher compared with that of baseline literature.ConclusionThe proposed registration method can obtain high-level feature information; thus,the registration accuracy is improved. Simultaneously,the loss function is built on the basis of the least-squares method,and the discriminator is also strengthened,which can enable the registration model to quickly converge during the training phase and then improve model stability and generalization ability. This method is suitable for medical image registration with large non-rigid deformation.关键词:medical image;non-rigid registration;generative adversarial network(GAN);residual-in-residual dense block(RRDB);least square loss;relative average GAN(RaGAN)33|31|0更新时间:2024-05-07
摘要:ObjectiveMedical image registration is an important research task in the field of medical image analysis,which is the basis of medical image fusion and medical image reconstruction. Conventional registration methods that build loss function based on normalized mutual information by using iterative gradient descent to achieve registration tend to be time consuming. The existing deep learning-based medical image registration methods have limitation in registering medical images with large non-rigid deformation,which cannot achieve high registration accuracy and have poor generalization ability. Thus,this paper proposes a method to register multi-modal medical images by combining residual-in-residual dense block (RRDB) with generative adversarial network (GAN).MethodFirst,RRDB are introduced to the standard generator network to extract high-level feature information from unpaired image pairs; thus,registration accuracy is improved. Then,a least-squares loss is used to substitute cross-entropy loss constructed by the logistic regression objective. The convergence condition of least-squares loss is strict and promotes the model to reach convergence at the optimal parameters,which can alleviate gradient disappearance and overfitting; thus,the robustness of model training is enhanced. In addition,relative average GAN (RaGAN) is embedded into the standard discriminator network,namely,adding a gradient penalty in the discriminator network,which reduce the error of discriminator to estimate the relative true and false probability between the fixed image and moving image. The enhanced discriminator can help the generator to learn clearer and texture information; therefore,the registration error can be decreased,and the registration accuracy can be stabilized.ResultThis registration model is trained and validated on DRIVE(digital retinal images for vessel extraction) dataset. Generalization performance tests are performed on Sunybrook Cardiac Dataset and Brain MRI Dataset. Compared with state-of-the-art registration methods,extensive experiments demonstrate that the proposed model achieves good registration results; both registration accuracy and generalization ability have been improved. Compared with the basic literature,the registration Dice values of retinal images,cardiac images,and brain images are improved by 3.3%,3.0%,and 1.5%,respectively. According to the stability verification experiment of the registration model,as the number of iterations augments,the Dice value of this paper gradually increases,and the change is more stable than that of baseline literature. The number of iterations in this paper is 80 000,whereas that of baseline literature is 10 000. This verification experiment shows that this registration model has been effectively improved in the training phase; not only the convergence speed is accelerated but also stability is higher compared with that of baseline literature.ConclusionThe proposed registration method can obtain high-level feature information; thus,the registration accuracy is improved. Simultaneously,the loss function is built on the basis of the least-squares method,and the discriminator is also strengthened,which can enable the registration model to quickly converge during the training phase and then improve model stability and generalization ability. This method is suitable for medical image registration with large non-rigid deformation.关键词:medical image;non-rigid registration;generative adversarial network(GAN);residual-in-residual dense block(RRDB);least square loss;relative average GAN(RaGAN)33|31|0更新时间:2024-05-07
Magnetic Resonance Image
-
 摘要:ObjectiveUltrasound is a main imaging method used for the diagnosis of thyroid diseases. It is convenient for the diagnosis of medical results through the real-time study of its internal anatomical structure. In computer vision,the segmentation of image tissue and organ is the pre background classification of the pixels in the image. The final segmentation image boundary is the combination of the target pixels. The research on medical image segmentation has received much attention,which is mainly divided into two ideas,where the first idea is to obtain the target area by analyzing the pixel value of a given image through computer vision technology. However,the generalization ability of the given image analysis is poor,and the segmentation effect is unremarkable because of the interference of random noise in the ultrasonic image. The second idea is to use deep learning for obtaining the target area through the background information before deep convolution classification. However,the target area may be insignificant using the depth learning model because of the complexity of tissue and organs,the evident surrounding tissues,and the lack of background information before the image,making the abstract features obtained by the depth network mostly the surrounding non target area and causing the segmentation effect of the original target unideal. A thyroid image is different in shape,size,and texture among different data sources. To solve the two problems,a thyroid ultrasound segmentation network based on frequency domain enhancement and local attention mechanism is proposed to solve the problem of random noise interference and insignificant target.MethodFirst,high and low pass filters are used to obtain the image information of high- and low-frequency bands,and the detail features of high frequency band and the edge feature of low frequency band are integrated to enhance the contrast of background and reduce the difference between images. Second,a local attention mechanism is used to adaptively activate the high- and low-dimensional feature information in accordance with the different information amounts of the features extracted by the network depth in the convolution network. This mechanmism can enhance the detailed information of low-dimensional features,weaken the attention to nontarget areas,enhance the global information of high-dimensional features,and weaken the interference of redundant information on the network,thereby enhancing the ability of background classification and nonsignificant target detection. Finally,a pyramid cascading hole is used,and convolution is utilized to obtain the feature information of different receptive fields and solve the problem of large image difference between data sources. In the training process,a mixed loss function is used to regress the network training effect,and pixel level loss (binary cross entropy) and image similarity loss (structural similarity) can better evaluate the segmentation prediction results. This paper uses the ResNet34 network,which is trained in advance to fine tune,to train the model of the network. The training set adopts the open data set of the network and selects approximately 3 500 images through the screening of appropriate images. During the training,one NVIDIA P100 graphics processing unit(GPU) server is used,the network training of approximately 10 epochs can achieve a better and stable effect,and the total training time is approximately 120 min.ResultExperimental results show that the accuracy of the proposed method is 0.989,the recall rate is 0.849,the specificity is 0.94,and the Dice coefficient is 0.812,which is better than the current methods of medical image segmentation network,such as U-Net and CE-Net network,and is more accurate and special in the effect of ultrasound thyroid image segmentation. A significant improvement is found in heterosexuality and is better than the evaluation result for the network using the same dataset,such as sumNet. At the same time,the ablation experiments show that the proposed modules have a certain improvement effect on ultrasound image segmentation.ConclusionThe proposed segmentation model combined with the advantages of deep learning model and traditional image processing model can better deal with ultrasound image random spots and improve the results of nonsignificant tissue segmentation.关键词:image segmentation;frequency domain analysis;attention mechanism;dilate convolution;ultrasound image66|176|3更新时间:2024-05-07
摘要:ObjectiveUltrasound is a main imaging method used for the diagnosis of thyroid diseases. It is convenient for the diagnosis of medical results through the real-time study of its internal anatomical structure. In computer vision,the segmentation of image tissue and organ is the pre background classification of the pixels in the image. The final segmentation image boundary is the combination of the target pixels. The research on medical image segmentation has received much attention,which is mainly divided into two ideas,where the first idea is to obtain the target area by analyzing the pixel value of a given image through computer vision technology. However,the generalization ability of the given image analysis is poor,and the segmentation effect is unremarkable because of the interference of random noise in the ultrasonic image. The second idea is to use deep learning for obtaining the target area through the background information before deep convolution classification. However,the target area may be insignificant using the depth learning model because of the complexity of tissue and organs,the evident surrounding tissues,and the lack of background information before the image,making the abstract features obtained by the depth network mostly the surrounding non target area and causing the segmentation effect of the original target unideal. A thyroid image is different in shape,size,and texture among different data sources. To solve the two problems,a thyroid ultrasound segmentation network based on frequency domain enhancement and local attention mechanism is proposed to solve the problem of random noise interference and insignificant target.MethodFirst,high and low pass filters are used to obtain the image information of high- and low-frequency bands,and the detail features of high frequency band and the edge feature of low frequency band are integrated to enhance the contrast of background and reduce the difference between images. Second,a local attention mechanism is used to adaptively activate the high- and low-dimensional feature information in accordance with the different information amounts of the features extracted by the network depth in the convolution network. This mechanmism can enhance the detailed information of low-dimensional features,weaken the attention to nontarget areas,enhance the global information of high-dimensional features,and weaken the interference of redundant information on the network,thereby enhancing the ability of background classification and nonsignificant target detection. Finally,a pyramid cascading hole is used,and convolution is utilized to obtain the feature information of different receptive fields and solve the problem of large image difference between data sources. In the training process,a mixed loss function is used to regress the network training effect,and pixel level loss (binary cross entropy) and image similarity loss (structural similarity) can better evaluate the segmentation prediction results. This paper uses the ResNet34 network,which is trained in advance to fine tune,to train the model of the network. The training set adopts the open data set of the network and selects approximately 3 500 images through the screening of appropriate images. During the training,one NVIDIA P100 graphics processing unit(GPU) server is used,the network training of approximately 10 epochs can achieve a better and stable effect,and the total training time is approximately 120 min.ResultExperimental results show that the accuracy of the proposed method is 0.989,the recall rate is 0.849,the specificity is 0.94,and the Dice coefficient is 0.812,which is better than the current methods of medical image segmentation network,such as U-Net and CE-Net network,and is more accurate and special in the effect of ultrasound thyroid image segmentation. A significant improvement is found in heterosexuality and is better than the evaluation result for the network using the same dataset,such as sumNet. At the same time,the ablation experiments show that the proposed modules have a certain improvement effect on ultrasound image segmentation.ConclusionThe proposed segmentation model combined with the advantages of deep learning model and traditional image processing model can better deal with ultrasound image random spots and improve the results of nonsignificant tissue segmentation.关键词:image segmentation;frequency domain analysis;attention mechanism;dilate convolution;ultrasound image66|176|3更新时间:2024-05-07 -
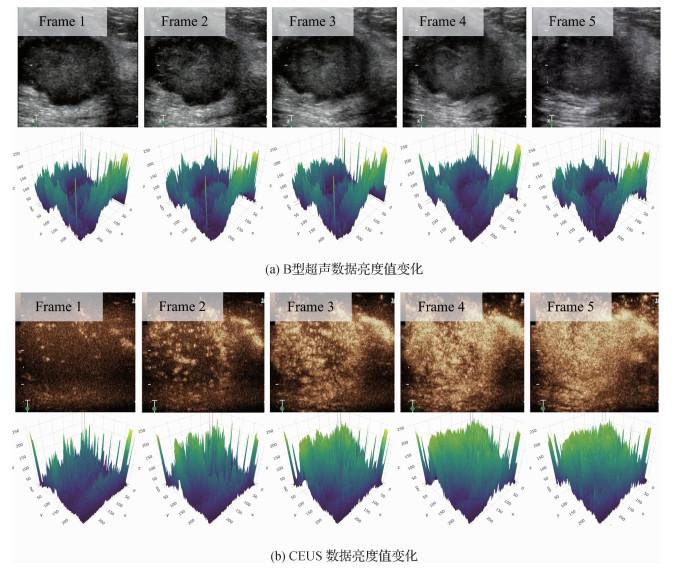 摘要:ObjectiveComputer-aided breast cancer diagnosis is a fundamental problem in the field of medical imaging. Correct diagnosis of breast cancer through deep learning can immensely improve the patients' survival rate. At present, most researchers only use B-mode ultrasound images as experimental data, but the limitation of B-mode ultrasound data makes it difficult to achieve a high classification accuracy. With the development of medical images, contrast-enhanced ultrasound (CEUS) video can provide accurate pathological information by observing the dynamic enhancement of the lesion area in temporal sequence. In view of the above ultrasound image problems, this paper proposes a network model that can comprehensively utilize B-mode ultrasound video data and CEUS video data to improve the classification accuracy.MethodFirst, a dual-branch model architecture is designed on the basis of the characteristics of two-stream structure and dual-modal data. One branch uses a frame of B-mode ultrasound video data and Resnet34 network model to extract pathological features. The other branch uses ultrasound contrast data and R (2+1) network model to extract temporal sequence information. Second, pathological multilabel pretraining is designed in this branch using 10 pathological information in CEUS video data because of the shortcoming of traditional video feature extraction. After the two-branch network, the characteristics of B-made ultrasound data and CEUS video data are obtained. We perform bilinear fusion on the obtained features to better integrate the features of B-mode ultrasound and CEUS. To extract pathological information and suppress irrelevant noise, the extracted and fused features from the two-branch network are processed using the attention mechanism to obtain the attention weight of the corresponding feature, and the corresponding weight is applied to the original feature. Weighted ultrasound and contrast features are obtained. Finally, the features obtained through the attention mechanism are bilinearly fused to obtain the final features.ResultThis article designed three experiments, where the first two experiments are pretraining on B-mode ultrasound and CEUS to verify the effectiveness of the proposed method and select the network with the strongest feature extraction ability for ultrasound data. In the B-mode ultrasound data pretraining experiment, the classic VGG(visual geometry group)16-BN(batch normalization), VGG19-BN, ResNet13, ResNet34, and ResNet50 networks were selected as the backbone network of the ultrasound branch for training to select the network with the strongest extraction ability for ultrasound images. The final classification results of each network are 74.2%, 75.6%, 80.5%, 81.0%, and 92.1%. Considering that the accuracy of the Resnet50 network in the test set is only 79.3%, which is relatively different from the accuracy of the training set and resulting in serious overfitting, the Resnet34 network is used as the backbone network of B-mode ultrasound data. In the pretraining experiment of the CEUS branch, the current mainstream P3D, R3D, CM3, and R (2+1) D convolutional networks are used as the backbone network of the CEUS branch for training. The final classification results of each network are 75.2%, 74.6 %, 74.1%, and 78.4%, and the R (2+1) D network with better results in the experiment is selected as the backbone network of the CEUS branch. Pretraining using pathological multilabels is designed in accordance with the medical field. The accuracy of the experiment combining the two data is improved by 6.5% compared with the use of B-mode ultrasound images alone and improved by 7.9% compared with the single-use CEUS video. At the same time, the proposed method achieves the highest accuracy in the use of bimodal data, which increases by 2.7% compared with the highest score.ConclusionThe proposed cooperative suppression network can process different modal data differently to extract the pathological features. On the one hand, multimodal data can certainly display the same lesion area from different angles, providing many pathological features for the classification model, thereby improving its classification accuracy. On the other hand, a proper fusion method is crucial because it can maximize the use of features and suppress noise.关键词:contrast-enhanced ultrasound(CEUS);double-branch network;bilinear fusion;collaborative attention;breast cancer classification23|22|1更新时间:2024-05-07
摘要:ObjectiveComputer-aided breast cancer diagnosis is a fundamental problem in the field of medical imaging. Correct diagnosis of breast cancer through deep learning can immensely improve the patients' survival rate. At present, most researchers only use B-mode ultrasound images as experimental data, but the limitation of B-mode ultrasound data makes it difficult to achieve a high classification accuracy. With the development of medical images, contrast-enhanced ultrasound (CEUS) video can provide accurate pathological information by observing the dynamic enhancement of the lesion area in temporal sequence. In view of the above ultrasound image problems, this paper proposes a network model that can comprehensively utilize B-mode ultrasound video data and CEUS video data to improve the classification accuracy.MethodFirst, a dual-branch model architecture is designed on the basis of the characteristics of two-stream structure and dual-modal data. One branch uses a frame of B-mode ultrasound video data and Resnet34 network model to extract pathological features. The other branch uses ultrasound contrast data and R (2+1) network model to extract temporal sequence information. Second, pathological multilabel pretraining is designed in this branch using 10 pathological information in CEUS video data because of the shortcoming of traditional video feature extraction. After the two-branch network, the characteristics of B-made ultrasound data and CEUS video data are obtained. We perform bilinear fusion on the obtained features to better integrate the features of B-mode ultrasound and CEUS. To extract pathological information and suppress irrelevant noise, the extracted and fused features from the two-branch network are processed using the attention mechanism to obtain the attention weight of the corresponding feature, and the corresponding weight is applied to the original feature. Weighted ultrasound and contrast features are obtained. Finally, the features obtained through the attention mechanism are bilinearly fused to obtain the final features.ResultThis article designed three experiments, where the first two experiments are pretraining on B-mode ultrasound and CEUS to verify the effectiveness of the proposed method and select the network with the strongest feature extraction ability for ultrasound data. In the B-mode ultrasound data pretraining experiment, the classic VGG(visual geometry group)16-BN(batch normalization), VGG19-BN, ResNet13, ResNet34, and ResNet50 networks were selected as the backbone network of the ultrasound branch for training to select the network with the strongest extraction ability for ultrasound images. The final classification results of each network are 74.2%, 75.6%, 80.5%, 81.0%, and 92.1%. Considering that the accuracy of the Resnet50 network in the test set is only 79.3%, which is relatively different from the accuracy of the training set and resulting in serious overfitting, the Resnet34 network is used as the backbone network of B-mode ultrasound data. In the pretraining experiment of the CEUS branch, the current mainstream P3D, R3D, CM3, and R (2+1) D convolutional networks are used as the backbone network of the CEUS branch for training. The final classification results of each network are 75.2%, 74.6 %, 74.1%, and 78.4%, and the R (2+1) D network with better results in the experiment is selected as the backbone network of the CEUS branch. Pretraining using pathological multilabels is designed in accordance with the medical field. The accuracy of the experiment combining the two data is improved by 6.5% compared with the use of B-mode ultrasound images alone and improved by 7.9% compared with the single-use CEUS video. At the same time, the proposed method achieves the highest accuracy in the use of bimodal data, which increases by 2.7% compared with the highest score.ConclusionThe proposed cooperative suppression network can process different modal data differently to extract the pathological features. On the one hand, multimodal data can certainly display the same lesion area from different angles, providing many pathological features for the classification model, thereby improving its classification accuracy. On the other hand, a proper fusion method is crucial because it can maximize the use of features and suppress noise.关键词:contrast-enhanced ultrasound(CEUS);double-branch network;bilinear fusion;collaborative attention;breast cancer classification23|22|1更新时间:2024-05-07 -
 摘要:ObjectiveBreast cancer has become one of the highest mortality rates in women. The incidence rate of breast cancer has been increasing in recent years. Early diagnosis of breast cancer is the key to treatment and control. Clinical data show that early diagnosis can reduce the mortality by 40%. Medical ultrasound has high accuracy, without trauma and radiation to the body, and requires extremely low cost. Therefore, ultrasound is often used as the first choice in medical diagnosis. Computer-aided diagnosis combines the powerful analysis and calculation ability of computers with medical image processing, thereby immensely improving the working efficiency and reducing the missed diagnosis and misdiagnosis. Deep learning has achieved excellent performance in various semantic segmentation tasks, and its accuracy is far beyond the traditional method. A common application of deep learning is focus segmentation. The focus area is the location of the lesion, which is usually part of the entire ultrasound image. Before diagnosis, the focus area is divided as the region of interest. On the one hand, it can reduce the amount of computation to improve the speed of the algorithm. On the other hand, it can reduce the background interference to improve the accuracy of the algorithm. The training of deep learning needs a large number of training samples with accurate labels. The precise location and segmentation of the focus area need to be manually labeled. Breast ultrasound results in low contrast and resolution, high speckle noise, and blurry boundary between tissues because of its imaging characteristics. These problems make accurate labeling extremely difficult. At present, breast ultrasound is the common method for experienced doctors to accurately label the focus area. However, this labor-intensive work aggravates the workload of doctors and seriously affects the diagnosis efficiency of doctors. Thus, studying the automatic location and segmentation of the focus area in ultrasound images is of great significance. Although the ultrasonic segmentation data set marked by doctors is uncleaned many times, many inaccurate marks are found in the data set, which are called mark noise. The training set will immensely affect the accuracy of network segmentation when it contains a certain amount of noise. To solve this problem, this paper proposes a dynamic noise index (DNI) and a novel loss function to achieve the segmentation of breast ultrasound tumor in noisy environment.MethodIn this paper, DNI is proposed to realize the detectability of noise. The DNI is then updated in real time during network training. At the end of each iteration, the DNI distribution of the entire training set is calculated. The detection of noise data is realized through the monitoring of data distribution. This paper proposes an improved loss function to enhance the performance of network noise detection. This loss function combines DNI to reduce the influence of noise on training back propagation. To verify the effectiveness of the algorithm, the DNI and the improved loss function are combined to form a noise tolerance framework. The framework can be applied to all types of segmented networks. The experiment is designed into two datasets, namely, open breast tumor segmentation dataset of Breast Ultrasound Images (DBUI) and Noisy Breast Ultrasound Image Dataset (NBUID). NBUID consists of 1 305 rough labeled breast tumor images. The validation and test sets consist of 500 exact annotation samples. Five hundred samples cover different nodule shapes (round, elliptical, irregular, lobulated, etc.), different nodule sizes, different nodule boundaries, different internal echoes, and different growth sites. 350 of them are test sets, and 150 are verification sets.ResultIn this paper, the noise tolerance framework is combined with three segmentation networks, namely, U-net, PSPNet, and FastFCN. The test results of DBUI30 show that the network with noise tolerance framework is 8%~12% higher than the original network. This paper constructs a data set of 1 805 images of breast ultrasound tumor segmentation. Combined with the noise tolerance framework, the performance of the network is improved by 4%~7%.ConclusionThe proposed DNI and segmentation algorithm can effectively suppress noise and achieve stable segmentation performance for training data sets with different noise ratios.关键词:breast ultrasound;breast tumor segmentation;noise annotation;deep learning;loss function35|19|0更新时间:2024-05-07
摘要:ObjectiveBreast cancer has become one of the highest mortality rates in women. The incidence rate of breast cancer has been increasing in recent years. Early diagnosis of breast cancer is the key to treatment and control. Clinical data show that early diagnosis can reduce the mortality by 40%. Medical ultrasound has high accuracy, without trauma and radiation to the body, and requires extremely low cost. Therefore, ultrasound is often used as the first choice in medical diagnosis. Computer-aided diagnosis combines the powerful analysis and calculation ability of computers with medical image processing, thereby immensely improving the working efficiency and reducing the missed diagnosis and misdiagnosis. Deep learning has achieved excellent performance in various semantic segmentation tasks, and its accuracy is far beyond the traditional method. A common application of deep learning is focus segmentation. The focus area is the location of the lesion, which is usually part of the entire ultrasound image. Before diagnosis, the focus area is divided as the region of interest. On the one hand, it can reduce the amount of computation to improve the speed of the algorithm. On the other hand, it can reduce the background interference to improve the accuracy of the algorithm. The training of deep learning needs a large number of training samples with accurate labels. The precise location and segmentation of the focus area need to be manually labeled. Breast ultrasound results in low contrast and resolution, high speckle noise, and blurry boundary between tissues because of its imaging characteristics. These problems make accurate labeling extremely difficult. At present, breast ultrasound is the common method for experienced doctors to accurately label the focus area. However, this labor-intensive work aggravates the workload of doctors and seriously affects the diagnosis efficiency of doctors. Thus, studying the automatic location and segmentation of the focus area in ultrasound images is of great significance. Although the ultrasonic segmentation data set marked by doctors is uncleaned many times, many inaccurate marks are found in the data set, which are called mark noise. The training set will immensely affect the accuracy of network segmentation when it contains a certain amount of noise. To solve this problem, this paper proposes a dynamic noise index (DNI) and a novel loss function to achieve the segmentation of breast ultrasound tumor in noisy environment.MethodIn this paper, DNI is proposed to realize the detectability of noise. The DNI is then updated in real time during network training. At the end of each iteration, the DNI distribution of the entire training set is calculated. The detection of noise data is realized through the monitoring of data distribution. This paper proposes an improved loss function to enhance the performance of network noise detection. This loss function combines DNI to reduce the influence of noise on training back propagation. To verify the effectiveness of the algorithm, the DNI and the improved loss function are combined to form a noise tolerance framework. The framework can be applied to all types of segmented networks. The experiment is designed into two datasets, namely, open breast tumor segmentation dataset of Breast Ultrasound Images (DBUI) and Noisy Breast Ultrasound Image Dataset (NBUID). NBUID consists of 1 305 rough labeled breast tumor images. The validation and test sets consist of 500 exact annotation samples. Five hundred samples cover different nodule shapes (round, elliptical, irregular, lobulated, etc.), different nodule sizes, different nodule boundaries, different internal echoes, and different growth sites. 350 of them are test sets, and 150 are verification sets.ResultIn this paper, the noise tolerance framework is combined with three segmentation networks, namely, U-net, PSPNet, and FastFCN. The test results of DBUI30 show that the network with noise tolerance framework is 8%~12% higher than the original network. This paper constructs a data set of 1 805 images of breast ultrasound tumor segmentation. Combined with the noise tolerance framework, the performance of the network is improved by 4%~7%.ConclusionThe proposed DNI and segmentation algorithm can effectively suppress noise and achieve stable segmentation performance for training data sets with different noise ratios.关键词:breast ultrasound;breast tumor segmentation;noise annotation;deep learning;loss function35|19|0更新时间:2024-05-07
Ultrasound Image
-
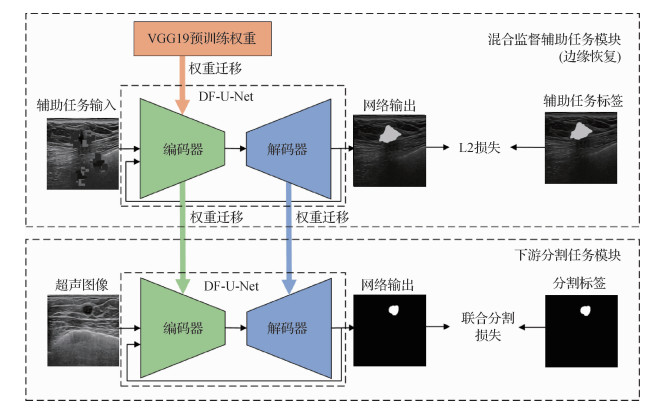 摘要:ObjectiveIn the clinical diagnosis and treatment of breast cancer, ultrasound imaging is widely used because of its real-time, non-radiation, and low cost. Automatic segmentation of breast lesions is a basic preprocessing step for computer-aided diagnosis and quantitative analysis of breast cancer. However, breast ultrasound segmentation is challenging. First, more noises and artifacts can be found in ultrasound images, and the boundary of the lesions is more ambiguous than the foreground in general segmentation tasks. Second, the size of the lesions in different sample images is different. In addition, benign and malignant lesions are quite different. The segmentation effect depends on the ability of the algorithm to understand the overall image. However, traditional methods rely on the characteristics of artificial design, which is difficult to deal with the noise and image structures. In recent years, excellent segmentation models such as U-Net are identified in the field of medical image segmentation. Many algorithms are based on U-Net, such as Auto-U-Net. Auto-U-Net uses the idea of iterative training to form a new input from the probability graph of model output and the original graph and sends them to the new U-Net model for training. However, the number of models needed in Auto-U-Net is the same as the total number of iterations, which leads to a complex training process and inefficient parameter utilization. The segmentation algorithm based on deep learning has a certain demand for data scale and annotation quality, whereas the professional requirements for accurate annotation of medical image data are high. Therefore, the number of samples cannot be guaranteed, resulting in the limited performance of the deep learning model. For the above mentioned challenges, in addition to transfer learning methods, using self-supervised learning to assist the training process is also a feasible solution. Considering that self-supervised learning emphasizes self-learning, this feature can deal with the problem of high cost in medical image field. Compared with the common transfer learning method, the advantage of self-supervised learning in the field of medical image lies in the stronger correlation between pretext task and target task. At present, in the field of medical image, the focus of research on self-supervised learning method is self-monitoring learning, whereas semantic segmentation is only a downstream task to evaluate self-supervised learning to features. In this process, the assistant task lacks the effective use of label information. Facing these limitations, this paper proposes a hybrid supervised dual-channel feedback U-Net (HSDF-U-Net) to improve the accuracy of breast ultrasound image segmentation.MethodHSDF-U-Net achieves hybrid supervised learning by integrating self-supervised learning and supervised segmentation and improves the accuracy of image segmentation by developing a dual channel feedback U-Net network. The algorithm designs the edge recovery task on the basis of the information in the segmentation label to enhance the correlation between the pretext task and the target task in self-supervised learning. The location information of contour pixels is extracted from the segmentation label. The images with ambiguous edge and the images with gray value close to the segmentation mask are obtained by using this information. They are used as input and label of deep learning to obtain the pre-training model with stronger ability to represent the edge of lesions and then transferred to the downstream image segmentation task. In addition, the feedback mechanism is introduced into U-Net to improve the performance of the model in the pretext edge restoration task and the downstream segmentation task. The mechanism is based on the general feed-forward convolutional neural network(CNN) and integrates the idea of weight sharing in the recurrent neural network. Through feeding back feature map, the prediction results are continuously refined. Therefore, we propose a dual channel feedback U-Net. The output probability map is fed back to the coding stage as the input of the encoder part probability channel. It forms a dual channel input together with ultrasonic image, which is encoded and fused separately before decoding. Consequently, the prediction results are continuously refined.ResultThe performance of HSDF-U-Net algorithm was evaluated on two open breast ultrasound image segmentation datasets. HSDF-U-Net segmented the image in Dataset B obtained sensitivity of 0.848 0, dice of 0.826 1, and the average symmetrical surface distance of 5.81. The sensitivity of 0.803 9, dice of 0.803 1, and the average symmetrical surface distance of 6.44 were obtained on Dataset breast ultrasound images(BUSI). The above mentioned results were improved compared with the typical deep learning segmentation algorithm.ConclusionIn this study, the proposed HSDF-U-Net improves the accuracy of breast ultrasound image segmentation, indicating potential application value.关键词:breast ultrasound image segmentation;deep learning;self-supervised learning;hybrid supervised learning;dual-channel feedback U-Net42|66|3更新时间:2024-05-07
摘要:ObjectiveIn the clinical diagnosis and treatment of breast cancer, ultrasound imaging is widely used because of its real-time, non-radiation, and low cost. Automatic segmentation of breast lesions is a basic preprocessing step for computer-aided diagnosis and quantitative analysis of breast cancer. However, breast ultrasound segmentation is challenging. First, more noises and artifacts can be found in ultrasound images, and the boundary of the lesions is more ambiguous than the foreground in general segmentation tasks. Second, the size of the lesions in different sample images is different. In addition, benign and malignant lesions are quite different. The segmentation effect depends on the ability of the algorithm to understand the overall image. However, traditional methods rely on the characteristics of artificial design, which is difficult to deal with the noise and image structures. In recent years, excellent segmentation models such as U-Net are identified in the field of medical image segmentation. Many algorithms are based on U-Net, such as Auto-U-Net. Auto-U-Net uses the idea of iterative training to form a new input from the probability graph of model output and the original graph and sends them to the new U-Net model for training. However, the number of models needed in Auto-U-Net is the same as the total number of iterations, which leads to a complex training process and inefficient parameter utilization. The segmentation algorithm based on deep learning has a certain demand for data scale and annotation quality, whereas the professional requirements for accurate annotation of medical image data are high. Therefore, the number of samples cannot be guaranteed, resulting in the limited performance of the deep learning model. For the above mentioned challenges, in addition to transfer learning methods, using self-supervised learning to assist the training process is also a feasible solution. Considering that self-supervised learning emphasizes self-learning, this feature can deal with the problem of high cost in medical image field. Compared with the common transfer learning method, the advantage of self-supervised learning in the field of medical image lies in the stronger correlation between pretext task and target task. At present, in the field of medical image, the focus of research on self-supervised learning method is self-monitoring learning, whereas semantic segmentation is only a downstream task to evaluate self-supervised learning to features. In this process, the assistant task lacks the effective use of label information. Facing these limitations, this paper proposes a hybrid supervised dual-channel feedback U-Net (HSDF-U-Net) to improve the accuracy of breast ultrasound image segmentation.MethodHSDF-U-Net achieves hybrid supervised learning by integrating self-supervised learning and supervised segmentation and improves the accuracy of image segmentation by developing a dual channel feedback U-Net network. The algorithm designs the edge recovery task on the basis of the information in the segmentation label to enhance the correlation between the pretext task and the target task in self-supervised learning. The location information of contour pixels is extracted from the segmentation label. The images with ambiguous edge and the images with gray value close to the segmentation mask are obtained by using this information. They are used as input and label of deep learning to obtain the pre-training model with stronger ability to represent the edge of lesions and then transferred to the downstream image segmentation task. In addition, the feedback mechanism is introduced into U-Net to improve the performance of the model in the pretext edge restoration task and the downstream segmentation task. The mechanism is based on the general feed-forward convolutional neural network(CNN) and integrates the idea of weight sharing in the recurrent neural network. Through feeding back feature map, the prediction results are continuously refined. Therefore, we propose a dual channel feedback U-Net. The output probability map is fed back to the coding stage as the input of the encoder part probability channel. It forms a dual channel input together with ultrasonic image, which is encoded and fused separately before decoding. Consequently, the prediction results are continuously refined.ResultThe performance of HSDF-U-Net algorithm was evaluated on two open breast ultrasound image segmentation datasets. HSDF-U-Net segmented the image in Dataset B obtained sensitivity of 0.848 0, dice of 0.826 1, and the average symmetrical surface distance of 5.81. The sensitivity of 0.803 9, dice of 0.803 1, and the average symmetrical surface distance of 6.44 were obtained on Dataset breast ultrasound images(BUSI). The above mentioned results were improved compared with the typical deep learning segmentation algorithm.ConclusionIn this study, the proposed HSDF-U-Net improves the accuracy of breast ultrasound image segmentation, indicating potential application value.关键词:breast ultrasound image segmentation;deep learning;self-supervised learning;hybrid supervised learning;dual-channel feedback U-Net42|66|3更新时间:2024-05-07
Ultrasound lmage
-
 摘要:ObjectiveThe task of classification of chest diseases is an important part in the field of medical image processing. Its purpose is to assist professional doctors to make accurate diagnosis and treatment through a computer automatic recognition system, which has important significance and role in clinical medicine. Medical image technologies play an important role in the recognition of chest diseases. They can provide doctors with clear internal textural and structural information. X-ray is a common and economical method for the diagnosis of various chest diseases. However, the main challenges in chest disease classification are that the location and size of the focus area of various diseases are different on X-ray films and the texture performance is diverse. The imbalance of sample data categories increases the task difficulty. Visual fatigue and other problems caused by long-term work, even for trained professional doctors, missed diagnosis, and misdiagnosis are also inevitable. Therefore, determining how to classify chest diseases automatically and accurately has become a popular topic in the field of medical image processing. At present, researchers mainly use the deep learning method to train a convolutional neural network to identify chest diseases automatically, but the overall recognition accuracy is inadequately high. Most methods cannot utilize the information on the feature channel. In view of the abovementioned challenges and the necessity to improve the recognition accuracy of current algorithms, this paper proposes a multilabel classification algorithm for chest diseases based on a dense squeeze-and-excitation network by using the deep learning method.MethodFirst, to utilize the information on the channel of the feature map, we add a squeeze-and-excitation module to DenseNet densely as the high-attention module of the feature channel; hence, the network can fully consider the feature information of each disease. In the process of network propagation, the network can enhance the transmission of useful information for the correct judgment of disease types and inhibit the transmission of useless information. Second, given that the parameters on the convolution kernel skeleton, i.e., the central cross position, are important and the ordinary square convolution kernel is random at the time of initialization, the ordinary square convolution kernel may be optimized in a direction that is not to strengthen the skeleton parameters. We use an asymmetric convolution block to replace the ordinary square convolution kernel, highlight the role of the parameters on the central cross position of the convolution kernel, and improve the feature extraction capability of the entire network. Lastly, considering that each disease sample data in the data set is relatively different and the learning difficulty also differs, this paper adopts the focal loss function to increase the loss weight of difficult-to-identify diseases and reduce the loss weight of easy-to-identify diseases. The network will pay more attention to the learning of the difficult-to-identify samples by using the focal loss function to improve the learning capability of the network for the types of difficult-to-identify diseases, improve the overall recognition accuracy of the network for chest diseases, and reduce the accuracy differences.ResultWe conduct experiments on the large multilabel dataset ChestX-ray14, which was published by National Institutes of Health(NIH). The experimental results show that the average recognition accuracy of the proposed algorithm for 14 chest diseases is higher than that of three existing classic and advanced algorithms, and the average area under ROC curve(AUC) value is 0.802. Meanwhile, the recognition accuracy of this algorithm is improved to some extent for some difficult-to-identify chest diseases. In accordance with the gradient-weighted class activation mapping algorithm, we can generate the heat map of the focus area and visualize the focus area of the chest disease classification algorithm model in the process of disease diagnosis. Comparison demonstrates that the positioning of the focus area in the hot map is basically consistent with the marking frame given by professional doctors. This finding proves the validity of the model, provides a visual explanation for disease diagnosis, and helps gain the trust of professional doctors for auxiliary diagnosis.ConclusionIn this paper, a multilabel classification algorithm based on a dense squeeze-and-excitation network is proposed for the recognition and classification of chest diseases. The experimental results show that our model is superior to several state-of-the-art approaches, with a higher average AUC value and stronger ability to diagnose some diseases that are difficult to identify. Our model is suitable for the classification of diseases and the recognition of chest X-ray images.关键词:chest X-ray;multi-label classification;densely connected network;disease diagnosis;medical image processing111|144|11更新时间:2024-05-07
摘要:ObjectiveThe task of classification of chest diseases is an important part in the field of medical image processing. Its purpose is to assist professional doctors to make accurate diagnosis and treatment through a computer automatic recognition system, which has important significance and role in clinical medicine. Medical image technologies play an important role in the recognition of chest diseases. They can provide doctors with clear internal textural and structural information. X-ray is a common and economical method for the diagnosis of various chest diseases. However, the main challenges in chest disease classification are that the location and size of the focus area of various diseases are different on X-ray films and the texture performance is diverse. The imbalance of sample data categories increases the task difficulty. Visual fatigue and other problems caused by long-term work, even for trained professional doctors, missed diagnosis, and misdiagnosis are also inevitable. Therefore, determining how to classify chest diseases automatically and accurately has become a popular topic in the field of medical image processing. At present, researchers mainly use the deep learning method to train a convolutional neural network to identify chest diseases automatically, but the overall recognition accuracy is inadequately high. Most methods cannot utilize the information on the feature channel. In view of the abovementioned challenges and the necessity to improve the recognition accuracy of current algorithms, this paper proposes a multilabel classification algorithm for chest diseases based on a dense squeeze-and-excitation network by using the deep learning method.MethodFirst, to utilize the information on the channel of the feature map, we add a squeeze-and-excitation module to DenseNet densely as the high-attention module of the feature channel; hence, the network can fully consider the feature information of each disease. In the process of network propagation, the network can enhance the transmission of useful information for the correct judgment of disease types and inhibit the transmission of useless information. Second, given that the parameters on the convolution kernel skeleton, i.e., the central cross position, are important and the ordinary square convolution kernel is random at the time of initialization, the ordinary square convolution kernel may be optimized in a direction that is not to strengthen the skeleton parameters. We use an asymmetric convolution block to replace the ordinary square convolution kernel, highlight the role of the parameters on the central cross position of the convolution kernel, and improve the feature extraction capability of the entire network. Lastly, considering that each disease sample data in the data set is relatively different and the learning difficulty also differs, this paper adopts the focal loss function to increase the loss weight of difficult-to-identify diseases and reduce the loss weight of easy-to-identify diseases. The network will pay more attention to the learning of the difficult-to-identify samples by using the focal loss function to improve the learning capability of the network for the types of difficult-to-identify diseases, improve the overall recognition accuracy of the network for chest diseases, and reduce the accuracy differences.ResultWe conduct experiments on the large multilabel dataset ChestX-ray14, which was published by National Institutes of Health(NIH). The experimental results show that the average recognition accuracy of the proposed algorithm for 14 chest diseases is higher than that of three existing classic and advanced algorithms, and the average area under ROC curve(AUC) value is 0.802. Meanwhile, the recognition accuracy of this algorithm is improved to some extent for some difficult-to-identify chest diseases. In accordance with the gradient-weighted class activation mapping algorithm, we can generate the heat map of the focus area and visualize the focus area of the chest disease classification algorithm model in the process of disease diagnosis. Comparison demonstrates that the positioning of the focus area in the hot map is basically consistent with the marking frame given by professional doctors. This finding proves the validity of the model, provides a visual explanation for disease diagnosis, and helps gain the trust of professional doctors for auxiliary diagnosis.ConclusionIn this paper, a multilabel classification algorithm based on a dense squeeze-and-excitation network is proposed for the recognition and classification of chest diseases. The experimental results show that our model is superior to several state-of-the-art approaches, with a higher average AUC value and stronger ability to diagnose some diseases that are difficult to identify. Our model is suitable for the classification of diseases and the recognition of chest X-ray images.关键词:chest X-ray;multi-label classification;densely connected network;disease diagnosis;medical image processing111|144|11更新时间:2024-05-07 -
 摘要:ObjectiveDental defect is a common problem in oral restoration treatment. For the symptoms of severe tooth loss, the traditional clinical treatment method is porcelain tooth restoration. This method primarily relies on the experience and technology of physicians and experts. Given the long time, this method leads to restoration precision is not easy to control. The current restoration method is to use the dental restoration computer aided design/computer aided manufacturing(CAD/CAM) system for restoration treatment. For example, in the CAD design of full-crown restorations, most methods use the standard tooth model in the standard database as the initial model to replace the defective tooth. A series of algorithms are used to properly deform the crown mesh model to reconstruct the target full-crown restoration model. This method uses the standard tooth model, lacks personalization, and requires a large number of man-machines in the design process. With regard to the interactive operation, the operator's technical experience is required, and the design contains a large degree of subjectivity. A personalized design method for the missing tooth crown based on the generative adversarial networks (HRGAN) is proposed to solve the personalized design of complex tooth morphology.MethodGiven the powerful processing ability of the HRGAN on the image, the three-dimensional model is reduced to a two-dimensional image, and the network can learn it. First, the depth distance of the tooth is calculated by using the multi-direction orthogonal projection method, and the high-resolution depth images of the occlusal surface, lingual side, and buccal side were constructed. The quality of the tooth image is optimized by the method of pixel value enhancement to subsequently use the generative confrontation network and better fit the morphological distribution of the crown. Image entropy describes the richness of the information carried by the image. Image entropy is used to evaluate the constructed depth image and construct a batch of learning data sets suitable for deep convolutional networks. Then, based on the multi-scale generative and discriminant models, the structure of the missing tooth restoration network model is constructed. The generative model generates the crown data by capturing the distribution of the crown data. The discriminant model is used to determine whether a sample is from expert design data rather than generated data. The two models are trained alternately and iteratively, playing with each other and constantly optimizing. The network merges a feature-matching loss on the basis of the discriminator to better fit the morphological features of the teeth and adds a perceptual loss to enhance network's ability to reconstruct the shape of the occlusal surface of the tooth. Finally, the point cloud data of the crown are calculated on the basis of the gray value and distance value mapping, and the rough registration is performed according to the information of the missing crown in the adjacent tooth. The iterative closest point algorithm is used to precisely register the generated crown point cloud with the original point cloud and achieve the reconstruction of the missing tooth model.ResultWe have constructed 500 sets of tooth depth map data, of which 400 sets are used as training sets and 100 sets are used as verification sets. The quality of the network-generated image is evaluated by the similarity between the tooth image synthesized from the HRGAN and the image obtained from the expert-designed tooth under experiments with different loss conditions. By calculating the peak signal to noise ratio(PSNR) and structrual similarity(SSIM) values of the two groups of images, the similarity between the images generated by the network and the image designed by the expert is improved after adding the loss of feature matching and perception, and the reconstructed occlusal surface of the crown has rich features of tooth pits and sulcus. The deviation analysis of the reconstructed occlusal surface and its corresponding expert design crown is carried out. The results show that compared with the CAD design method and the generative model-based design method, the standard deviation of the crown shape generated by the method in this paper is 21.2% and 7% lower than the former two methods, and the root mean square(RMS) value is 43.8% and 9.8% lower, respectively, and the shape of the occlusal surface of the crown is the closest to the shape of the expert-designed crown.ConclusionThe experimental results show that the tooth design method based on the high-resolution generation network proposed in this paper can effectively complete the morphological design of the missing tooth, and the designed crown has enough natural tooth anatomical morphological characteristics.关键词:full crown restoration;generative adversarial network(GAN);image synthesis;point cloud registration;tooth shape design69|77|2更新时间:2024-05-07
摘要:ObjectiveDental defect is a common problem in oral restoration treatment. For the symptoms of severe tooth loss, the traditional clinical treatment method is porcelain tooth restoration. This method primarily relies on the experience and technology of physicians and experts. Given the long time, this method leads to restoration precision is not easy to control. The current restoration method is to use the dental restoration computer aided design/computer aided manufacturing(CAD/CAM) system for restoration treatment. For example, in the CAD design of full-crown restorations, most methods use the standard tooth model in the standard database as the initial model to replace the defective tooth. A series of algorithms are used to properly deform the crown mesh model to reconstruct the target full-crown restoration model. This method uses the standard tooth model, lacks personalization, and requires a large number of man-machines in the design process. With regard to the interactive operation, the operator's technical experience is required, and the design contains a large degree of subjectivity. A personalized design method for the missing tooth crown based on the generative adversarial networks (HRGAN) is proposed to solve the personalized design of complex tooth morphology.MethodGiven the powerful processing ability of the HRGAN on the image, the three-dimensional model is reduced to a two-dimensional image, and the network can learn it. First, the depth distance of the tooth is calculated by using the multi-direction orthogonal projection method, and the high-resolution depth images of the occlusal surface, lingual side, and buccal side were constructed. The quality of the tooth image is optimized by the method of pixel value enhancement to subsequently use the generative confrontation network and better fit the morphological distribution of the crown. Image entropy describes the richness of the information carried by the image. Image entropy is used to evaluate the constructed depth image and construct a batch of learning data sets suitable for deep convolutional networks. Then, based on the multi-scale generative and discriminant models, the structure of the missing tooth restoration network model is constructed. The generative model generates the crown data by capturing the distribution of the crown data. The discriminant model is used to determine whether a sample is from expert design data rather than generated data. The two models are trained alternately and iteratively, playing with each other and constantly optimizing. The network merges a feature-matching loss on the basis of the discriminator to better fit the morphological features of the teeth and adds a perceptual loss to enhance network's ability to reconstruct the shape of the occlusal surface of the tooth. Finally, the point cloud data of the crown are calculated on the basis of the gray value and distance value mapping, and the rough registration is performed according to the information of the missing crown in the adjacent tooth. The iterative closest point algorithm is used to precisely register the generated crown point cloud with the original point cloud and achieve the reconstruction of the missing tooth model.ResultWe have constructed 500 sets of tooth depth map data, of which 400 sets are used as training sets and 100 sets are used as verification sets. The quality of the network-generated image is evaluated by the similarity between the tooth image synthesized from the HRGAN and the image obtained from the expert-designed tooth under experiments with different loss conditions. By calculating the peak signal to noise ratio(PSNR) and structrual similarity(SSIM) values of the two groups of images, the similarity between the images generated by the network and the image designed by the expert is improved after adding the loss of feature matching and perception, and the reconstructed occlusal surface of the crown has rich features of tooth pits and sulcus. The deviation analysis of the reconstructed occlusal surface and its corresponding expert design crown is carried out. The results show that compared with the CAD design method and the generative model-based design method, the standard deviation of the crown shape generated by the method in this paper is 21.2% and 7% lower than the former two methods, and the root mean square(RMS) value is 43.8% and 9.8% lower, respectively, and the shape of the occlusal surface of the crown is the closest to the shape of the expert-designed crown.ConclusionThe experimental results show that the tooth design method based on the high-resolution generation network proposed in this paper can effectively complete the morphological design of the missing tooth, and the designed crown has enough natural tooth anatomical morphological characteristics.关键词:full crown restoration;generative adversarial network(GAN);image synthesis;point cloud registration;tooth shape design69|77|2更新时间:2024-05-07 -
 摘要:ObjectiveWearable devices are expanding in terms of electrocardiograms (ECG) because they can portably monitor the heart condition of a human for a long time. The key for wearable devices to monitor heart conditions in real time is to be able to process the collected ECG data automatically. Therefore, an efficient and reliable classification algorithm for ECG data from wearable devices should be designed. Many ECG classification algorithms have been proposed in recent years. They are mainly divided into two categories: one is based on handcrafted features, and the other is based on deep learning. Classification methods based on artificial features need to design various features manually, and many useful features are often ignored. The deep learning method can automatically extract features, and it has achieved good performance in image classification, object recognition, and natural language processing. However, no ECG data set from wearable devices is publicly available. Most ECG analysis algorithms are aimed at ECG data collected using hospital equipment. The ECG signal collected using wearable devices is susceptible to various interferences due to the body movement of people and changes in the surrounding environment, resulting in various noises in the signal. Therefore, many methods that perform well in ECG data sets collected using hospital equipment cannot achieve accurate classification of ECG data from wearable devices. In this study, we used IREALCARE 2.0 flexible cardiac monitor patch as the wearable device to collect ECG signals and make ECG data sets. It has the characteristics of small size, lightweight, and accurate and reliable measurement. The ECG data set came from 38 subjects and mainly included five types of heartbeats: normal, ventricular premature beat (VPB), supraventricular premature beat (SPB), atrial fibrillation (AF), and interference. In accordance with the characteristics of ECG data from wearable devices, such as considerable interference and large amount of data, this study proposed a deep convolutional neural network (CNN) named time-spatial CNN (TSCNN) for the automatic classification and analysis of ECG signals from wearable devices.MethodTSCNN consisted of seven convolutional layers, four pooling layers, and one fully connected layer. It also comprised a convolution-pooling block, three convolution-convolution-pooling blocks, and a classification layer. First, the original long-term ECG signals were divided into separate heartbeats. We regarded the 600 data points around R-wave coordinates as one heartbeat, combined them with filtered heartbeat data of different frequency bands into 10 channels of data, and input them into TSCNN. Second, we applied convolution over time and spatial filtering for each heartbeat, which could effectively expand the receptive field of the network to extract abundant features. Lastly, cascaded convolution with a small convolution kernel was applied to improve classification performance and reduce the amount of network parameters and computation. In accordance with the input data size of each layer, we adopted three convolution kernels of different sizes. The convolution kernels in the three convolution-convolution-pooling blocks were 10×1, 5×1, and 3×1 in sequence with stride 1×1. The fully connected layer was used as the classification layer. Some regularized methods, such as dropout and batch normalization, were adopted to avoid overfitting. The batch normalization layer was placed after the convolutional layer and before the nonlinear layer, which made the input value of the nonlinear transformation function fall into a region sensitive to the input, to avoid the problem of gradient disappearance. Dropout randomly set some hidden layer nodes to zero during each iteration, which could reduce the interaction among hidden layer nodes.ResultThe proposed method was evaluated on the ECG data set produced in this paper and compared with four other ECG classification algorithms: CNN, recurrent neural network, 1D CNN (1-DCNN), and dense convolutional network (DCN). We used 10 records containing 138 853 heartbeats as the training set, and the test data were collected from 28 subjects, which contained 241 896 heartbeats. Results showed that the method in this paper achieved an overall accuracy of 91.16%, and the accuracies of normal, VPB, SPB, AF, and interference were 89.17%, 92.99%, 62.03%, 91.56%, and 94.89%, respectively. Comparative experiments indicated that our method was better than the four other methods in overall accuracy and the accuracies of normal, VPB, SPB, and AF and achieved a great improvement. Compared with 1-DCNN, the proposed method demonstrated increased overall accuracy by 7.25% and increased accuracies of normal, VPB, SPB, and AF by 18.13%, 12.67%, 9.29%, and 23.96%, respectively. Compared with DCN, the proposed method exhibited increased overall accuracy by 5.88% and increased accuracies of normal, VPB, SPB, and AF by 16.88%, 8.41%, 11.73%, and 20.32%, respectively. In addition, the classification results of our method were balanced, and the recognition capability for each class was similar. We also conducted a comparative experiment with and without spatial filtering. The experimental results demonstrated that spatial filtering could improve the classification performance of the network, and the overall accuracy increased from 88.17% to 91.16%. We further trained and evaluated deep networks that use cascaded convolution with a small convolution kernel and shallow networks that do not use it. The comparative experiment results showed that using cascaded convolution with a small convolution kernel could improve the classification capability of the network, in which the overall accuracy was increased by 1.56%; it also effectively reduced the amount of network parameters and calculations, thus increasing the processing speed.ConclusionIn this study, we proposed TSCNN to classify ECG signals from wearable devices. The experimental results indicated that the proposed method can achieve good classification performance for ECG data from wearable devices and effectively monitor whether the wearer has abnormal ECG.关键词:wearable devices;wearable ECG dataset;cardiac monitoring;convolutional neural network(CNN);spatial filtering36|39|1更新时间:2024-05-07
摘要:ObjectiveWearable devices are expanding in terms of electrocardiograms (ECG) because they can portably monitor the heart condition of a human for a long time. The key for wearable devices to monitor heart conditions in real time is to be able to process the collected ECG data automatically. Therefore, an efficient and reliable classification algorithm for ECG data from wearable devices should be designed. Many ECG classification algorithms have been proposed in recent years. They are mainly divided into two categories: one is based on handcrafted features, and the other is based on deep learning. Classification methods based on artificial features need to design various features manually, and many useful features are often ignored. The deep learning method can automatically extract features, and it has achieved good performance in image classification, object recognition, and natural language processing. However, no ECG data set from wearable devices is publicly available. Most ECG analysis algorithms are aimed at ECG data collected using hospital equipment. The ECG signal collected using wearable devices is susceptible to various interferences due to the body movement of people and changes in the surrounding environment, resulting in various noises in the signal. Therefore, many methods that perform well in ECG data sets collected using hospital equipment cannot achieve accurate classification of ECG data from wearable devices. In this study, we used IREALCARE 2.0 flexible cardiac monitor patch as the wearable device to collect ECG signals and make ECG data sets. It has the characteristics of small size, lightweight, and accurate and reliable measurement. The ECG data set came from 38 subjects and mainly included five types of heartbeats: normal, ventricular premature beat (VPB), supraventricular premature beat (SPB), atrial fibrillation (AF), and interference. In accordance with the characteristics of ECG data from wearable devices, such as considerable interference and large amount of data, this study proposed a deep convolutional neural network (CNN) named time-spatial CNN (TSCNN) for the automatic classification and analysis of ECG signals from wearable devices.MethodTSCNN consisted of seven convolutional layers, four pooling layers, and one fully connected layer. It also comprised a convolution-pooling block, three convolution-convolution-pooling blocks, and a classification layer. First, the original long-term ECG signals were divided into separate heartbeats. We regarded the 600 data points around R-wave coordinates as one heartbeat, combined them with filtered heartbeat data of different frequency bands into 10 channels of data, and input them into TSCNN. Second, we applied convolution over time and spatial filtering for each heartbeat, which could effectively expand the receptive field of the network to extract abundant features. Lastly, cascaded convolution with a small convolution kernel was applied to improve classification performance and reduce the amount of network parameters and computation. In accordance with the input data size of each layer, we adopted three convolution kernels of different sizes. The convolution kernels in the three convolution-convolution-pooling blocks were 10×1, 5×1, and 3×1 in sequence with stride 1×1. The fully connected layer was used as the classification layer. Some regularized methods, such as dropout and batch normalization, were adopted to avoid overfitting. The batch normalization layer was placed after the convolutional layer and before the nonlinear layer, which made the input value of the nonlinear transformation function fall into a region sensitive to the input, to avoid the problem of gradient disappearance. Dropout randomly set some hidden layer nodes to zero during each iteration, which could reduce the interaction among hidden layer nodes.ResultThe proposed method was evaluated on the ECG data set produced in this paper and compared with four other ECG classification algorithms: CNN, recurrent neural network, 1D CNN (1-DCNN), and dense convolutional network (DCN). We used 10 records containing 138 853 heartbeats as the training set, and the test data were collected from 28 subjects, which contained 241 896 heartbeats. Results showed that the method in this paper achieved an overall accuracy of 91.16%, and the accuracies of normal, VPB, SPB, AF, and interference were 89.17%, 92.99%, 62.03%, 91.56%, and 94.89%, respectively. Comparative experiments indicated that our method was better than the four other methods in overall accuracy and the accuracies of normal, VPB, SPB, and AF and achieved a great improvement. Compared with 1-DCNN, the proposed method demonstrated increased overall accuracy by 7.25% and increased accuracies of normal, VPB, SPB, and AF by 18.13%, 12.67%, 9.29%, and 23.96%, respectively. Compared with DCN, the proposed method exhibited increased overall accuracy by 5.88% and increased accuracies of normal, VPB, SPB, and AF by 16.88%, 8.41%, 11.73%, and 20.32%, respectively. In addition, the classification results of our method were balanced, and the recognition capability for each class was similar. We also conducted a comparative experiment with and without spatial filtering. The experimental results demonstrated that spatial filtering could improve the classification performance of the network, and the overall accuracy increased from 88.17% to 91.16%. We further trained and evaluated deep networks that use cascaded convolution with a small convolution kernel and shallow networks that do not use it. The comparative experiment results showed that using cascaded convolution with a small convolution kernel could improve the classification capability of the network, in which the overall accuracy was increased by 1.56%; it also effectively reduced the amount of network parameters and calculations, thus increasing the processing speed.ConclusionIn this study, we proposed TSCNN to classify ECG signals from wearable devices. The experimental results indicated that the proposed method can achieve good classification performance for ECG data from wearable devices and effectively monitor whether the wearer has abnormal ECG.关键词:wearable devices;wearable ECG dataset;cardiac monitoring;convolutional neural network(CNN);spatial filtering36|39|1更新时间:2024-05-07
Research and Application
0

