|
|
|
发布时间: 2022-06-16 |
数据挖掘和信息交互 |
|
|
|
收稿日期: 2022-02-27; 修回日期: 2022-03-03; 预印本日期: 2022-03-10
基金项目: 科技创新2030-“新一代人工智能”重大项目(2020AAA0109502);国家自然科学基金项目(U1809204,61525106,61427807,61701436,81871428)
作者简介:
叶慧慧,1988年生,女,讲师,主要研究方向为定量磁共振成像。E-mail: yehuihui@zju.edu.cn
何宏建,通信作者,男,副教授,主要研究方向为磁共振成像。E-mail:hhezju@zju.edu.cn 方静宛,女,博士后,主要研究方向为PET脑功能成像。E-mail:fangjingwan@zju.edu.cn 童琪琦,女,博士后,主要研究方向为多中心磁共振扩散成像融合校正、磁共振成像质量控制。E-mail:tongqq@zhejianglab.com 周子涵,女,博士研究生,主要研究方向为髓鞘磁共振成像。E-mail:zihan_zhou@zju.edu.cn 刘华锋,男,教授,主要研究方向为生物医学成像。E-mail: liuhf@zju.edu.cn *通信作者: 何宏建 hhezju@zju.edu.cn
中图法分类号: R-331
文献标识码: A
文章编号: 1006-8961(2022)06-1944-12
|
摘要
现代医学成像技术是脑科学研究和脑疾病诊断的利器,不同模态的成像技术提供不同的信息可协同表征脑部结构和功能。其中定量成像技术着眼于和生理、物理相关的内在参量,旨在提供更精准的信息。本文以正电子发射扫描成像(positron emission tomography,PET)和磁共振成像(magnetic resonance imaging,MRI)两种生物医学成像模态为例, 针对性地讨论它们在定量刻画大脑微观结构和功能领域的发展状况,目前尚存的关键技术问题和未来的可能发展方向。围绕定量MRI,从表观参数定量开始,介绍其中的单参数定量的现状和不足,以及目前多参数同时定量的发展动态;围绕微观参数定量,介绍针对髓鞘成像的两大方法,包括多组分T2定量和基于超短回波时间髓鞘直接成像,介绍磁共振定量成像特别是磁共振扩散成像的可比较性和可重复性研究。围绕定量PET,从最广泛的代谢动力学模型——房室模型开始介绍,对生理参数与示踪剂摄取量的关系进行了详细描述,展开到定量的误差来源包括模型选择、图像质量以及输入函数测量误差3个方面进行分析,介绍最新进展包括硬件设备、图像重建方法以及定量分析方法。最后对MRI定量、PET定量以及PET/MRI定量领域进行了展望。
关键词
多模态成像; 定量磁共振成像(MRI); 定量正电子发射扫描成像(PET); 多参数同时定量; 髓鞘水成分定量; 多中心融合; 房室模型
Abstract
Advanced medical imaging technology facilitates human brain recognition and its disease diagnosis research like positron emission tomography (PET) and magnetic resonance imaging (MRI). The changes of structure, function, metabolism, and signaling pathways yield richer multimodal image data for disease diagnosis research. Traditional clinical imaging techniques are mostly based on qualitative interpretation. The signal intensity of acquired images are differentiated for normal tissues, resulting in uncertainty in image contrast due to the microscopic scaled structure and function changes in tissue disease pathology. It is an effective way to obtain accurate and reliable detection of lesion features while the tissue contrast changes intensively higher than the noise level. In comparison to qualitative medical imaging, the current measurement focuses on physiology and physics related parameters to generate its quantitative parameter map. Quantitative parameters have their own physical units in common and their quantitative values reflect the physiological and physical information of the object mathematically. Quantitative measurement of tissue is essential to physiopathological modeling. The relationship between image nuances and pathology, realize in-depth clinical data mining for accurate diagnosis based on the integration of effective model analysis. The quantitative integration of cross-modalities and multiple imaging mechanisms medical imaging has been developed in brain tumors and neuropsychiatric diseases. While quantitative imaging technology is challenging in clinical settings, no matter due to its long acquisition time or its different image presentation. The common quantitative PET and MRI measurements are based on data fitting from multiple measurement. The multiple measurements are time consuming and costly. The modeling and simulation of micro-physiological systems still need to be continuously developed and improved, including the development from static models to dynamic models. Our research review and discuss the key technical issues and development of existing quantitative imaging technologies for human brain microstructure and physiological function indicators detection through PET and MRI methods. The clinical applications and future directions are introduced as well. Specifically, we focus on the establishment of quantitative models, the measurement of quantitative parameters and imaging methods, the influencing factors in the measurement, and the clinical application of related technologies. First, the review of quantitative MRI is based on the current situation and deficiencies of single-parameter quantification and the development trendency of simultaneous multi-parameter quantification. Then, it introduces two methods of myelin imaging based on the quantification of microscopic parameters, including multicomponent T2 quantification and ultrashort echo based myelin imaging. An introduction to the comparability and reproducibility of magnetic resonance quantitative imaging is followed on, especially magnetic resonance diffusion imaging. Second, the review of quantitative PET is based on the most extensive metabolic kinetic model-the compartment model. To extend quantitative error sources like model option, image quality, and input functions, the relationship between physiological parameters and tracer uptake is clarified and three aspects of measurement error are analyzed in detail. The latest development is reviewed based on hardware equipment, image reconstruction methods and quantitative analysis methods. The future MRI quantification, PET quantification and PET/MRI quantification are briefly predicted further.
Key words
multi-modal imaging; quantitative magnetic resonance imaging (MRI); quantitative positron emission tomography (PET); simultaneous multi-parameter quantification; myelin water faction quantification; multi-center fusion; compartment model
0 引言
现代医学成像技术是脑科学研究和脑疾病诊断的利器。以正电子发射扫描成像(positron emission tomography, PET)和磁共振成像(magnetic resonance imaging,MRI)两种重要技术为例,它们可以提供毫米尺度分辨率下大脑组织在细胞甚至分子水平有关的结构、功能、代谢和信号通路等变化信息,为疾病的诊断和研究提供了丰富的多模态图像数据。传统临床影像技术多以基于图像灰度值的定性比较为主。受微观尺度的疾病病理引起组织的结构和功能改变影响,医学图像局部信号强度将异于正常组织,导致图像对比度的显著变化。如果组织对比度变化显著高于噪声水平,就可以准确可靠地检测病灶特征。
近年来,定量成像技术发展成为新的热点。与传统医学影像不同,定量成像的测量目标超越灰度值变化,而着眼于与生理、物理相关参量的具体描述,并得到相应定量参数图。一般地,定量参数都有明确的物理单位;其量化数值是反映检测对象生理和物理信息的数学响应。临床常用的定性图像是检测对象的定量描述在给定成像参数条件下的特例表示。理论上,定量成像的结果仅与组织特性有关,因而可以跨设备、跨时间直接比较。此外,组织的定量测量是生理病理建模的重要基础。结合有效的模型分析,研究人员可以深入地揭示图像细微差别与病理的联系,实现对临床数据的深度挖掘,更好地为精准诊断目标服务。随着相关技术不断发展和成熟,融合跨模态和多种成像机制的定量医学成像已经广泛应用于脑肿瘤、神经精神疾病等重大脑疾病中的研究中,为深入理解和表征病理变化、实现疾病的纵向对比和预测带来新的契机。
现阶段,定量成像技术的发展还存在诸多技术难点。一方面,常见的PET和MRI定量测量建立在数据拟合的基础上。由此造成的多次测量导致成像过程复杂且成像时间冗长。成像系统硬件和成像原理、重建算法的改进,促成了快速成像领域的革命性突破,给定量成像技术的快速发展带来的巨大的契机。另一方面,人体是一个非常复杂的存在,微观生理系统的建模和仿真仍需不断发展和完善,包括从静态模型到动态模型的发展、从简单到复杂模型的发展。本文结合已有的工作经验,以PET和MRI两种关键定量脑成像系统为例,针对性地总结和讨论面向人脑微观结构和生理功能指标检测目标的定量成像技术发展现状、存在的关键技术问题和未来的可能发展方向。在具体描述中,本文将围绕定量模型建立、定量参数的测量及成像方法、测量中的影响因素、以及相关技术的应用案例等几个方面展开介绍。
1 定量磁共振成像
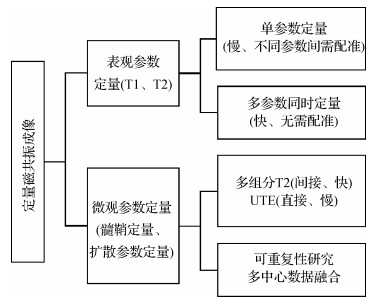
磁共振成像通常以体内氢质子作为对比剂,以组织不同的参数包括质子密度(proton density,PD)、纵向弛豫时间(T1)和横向弛豫时间(T2)的差异形成对比度。定量磁共振成像旨在提供更特异性的定量参数以利于疾病的表征。定量磁共振成像的参数分为表观参数和微观参数,其中表观参数包括PD、T1和T2等,微观参数包括髓鞘水分数、髓鞘组织分数、扩散系数和各向异性分数等,图 1是本文对定量磁共振成像方法和优劣的总结。
1.1 表观参数多参数同时定量
传统T1、T2弛豫参数定量是通过2维反转恢复自旋回波(inversion recovery spin echo, IR-SE)和多回波自旋回波(multi echo spin echo, ME-SE)信号进行指数拟合得到(Bernstein等,2004)。但这类方法非常耗时,难以在临床实践中使用。后来的Look-Locker方法(Look和Locker,1970)在IR脉冲后施加一系列小翻转角(flip angle, FA) 脉冲,并在每个脉冲后读出,实现了T1的加速定量。另一种更广泛使用的方法是基于梯度回波序列的T1定量方法(driven equilibrium single pulse observation of T1, DESPOT1)(Homer和Beevers,1985),可实现快速3D高分辨率T1定量。该方法通过两次以上不同FA的FLASH扫描,并将稳态的信号模型拟合得到T1定量参数。在此基础上,Deoni等人(2003)还开发了基于TrueFISP的DESPOT2用于T2定量。由于TrueFISP序列混合了T1和T2对比,通过至少两次不同FA扫描,先使用DESPOT1得到T1信息,进而拟合得到T2。
上述定量方法的共同点是一个序列完成多组不同扫描,但仅拟合一个定量参数。因此总体的测量效率较低。新近研究的热点关注在单个序列同时定量多个参量,即多参数同时定量。这一方面提高了采集的效率,同时还避免了序列间运动带来的配准困难。IR-TrueFISP(IR-true fast imaging with steady state precession)(Schmitt等,2004)方法通过一个序列的时间信号扫描,推导其解析表达式后可直接计算T1、T2和PD。此外,基于饱和恢复快速自旋回波的多参数定量序列(quantification of relaxation times and proton density by multiecho acquisition of a saturation-recovery using turbo spin-echo readout, QRAPMASTER(Warntjes等,2008)是基于饱和脉冲准备的自旋回波序列,通过多个准备脉冲的激发和不同层面的顺序错位实现不同T1加权,通过多个180°脉冲实现多个回波不同T2加权信号的获取,由此分别获得T1和T2定量。磁共振指纹成像(magnetic resonance fingerprinting, MRF)(Ma等,2013)突破了传统思路,广泛地变换IR-TrueFISP序列的基础扫描参数(如FA、TR等),引入时间信号的动态性,再使用字典匹配的方法同时获得多个定量参数。在MRF这一框架下,序列的设计更为灵活和多样,可实现额外引入其他参数,如B1+(Cloos等,2016)、T2*(Rieger等,2017)、灌注系数(Christen等,2014;Su等,2017)、CEST(chemical exchamge saturation transfer)(Zhou等,2018;Cohen等,2018)等进行更多参数的同时定量测量。并行成像、压缩感知成像和多层同时成像(simultaneous multi-slice, SMS)等通用加速方法可以实现进一步加速MRF(Liao等,2017;Ye等,2016, 2017)。近期研究已经在2 min左右实现全脑1 mm各向同性分辨率的T1、T2定量(Cao等,2019, 2022)。
为了区分病理组织和正常组织,T1和T2等同时定量在临床中有重要的应用价值,如区分肿瘤病灶、癫痫病灶、中风病灶和多发性硬化病灶等。该方向已成为一种技术发展趋势。例如,使用多任务(multitasking)方法同时全脑定量T1、T2和T1p,在多发性硬化症上展现了比单独一种定量参量更佳的鉴别诊断效果(Ma等,2021)。在颞叶内侧癫痫患者的海马硬化灶检测问题上,基于MRF技术的T1、T2定量参数比传统T1和T2定性成像具有更高的诊断准确性(Liao等,2018)。
1.2 微观髓鞘定量方法
大脑由多种不同类型的细胞组成,包括数以百亿单位的神经胶质细胞、皮层神经元和与之相连的神经纤维。以白质为例,微观尺度下轴突是纤维传导束的主要结构,它又被髓鞘包裹保护,并完成信号的快速传导。髓鞘双分子层之间的空间充满了水,通常称为髓鞘水。建立在毫米尺度的脑白质磁共振信号的常用模型包含细胞内水(intra-cellular water)、细胞外水(extra-cellular water)和髓鞘水3大部分。在3T磁场条件下,髓鞘水的T2弛豫时间约为10 ms,而髓鞘本身的T2更短,小于1 ms。细胞内/外水的T2弛豫时间为30~60 ms。相对比而言,脑脊液的T2弛豫时间大于1 s。因此,在微结构定量研究模型里,白质体素的信号应由多个不同组分T2信号衰减共同加权而成。基于定量磁共振成像和多组分模型的髓鞘水成像,是测量髓鞘水相对含量的重要方法,可以深入解析白质组织的微结构。相关技术已应用于创伤性轴突损伤(Choi等,2019)、衰老(Faizy等,2020)和多发性硬化(Baranovicova等,2016)等疾病的研究中。
髓鞘水成像得到的髓鞘水分数与独立测量的髓磷脂含量间有很强的相关性(Björk等,2016;Nagtegaal等,2020),但仍然是一种间接测量白质微结构的方法。由于髓鞘大分子的T2非常短,信号衰减极快,常规MRI方法无法检测。超短回波时间序列(ultrashort echo time, UTE)(Wilhelm等,2012)是直接检测这些信号的可行途径。例如,UTE-T2*成像采用一系列不同回波时间(包括0.5 ms或更短TE)的多幅图像进行指数拟合,可以测得与髓鞘大分子相关的超短T2*信号(Shao等,2016), 反转恢复超短回波时间技术(inversion recovery prepared UTE, IR-UTE)抑制了常规组织的信号,可以减少信号的干扰(Sheth等,2016)。但翻转时间TI的选择对定量结果准确性影响深刻。一种改进的技术方案是采用多个激发脉冲,更有效地抑制住长T2*组织(Ma等,2020a, b)。然而,该技术的成像时间偏长。一次全脑采集需要将近1 h,限制住了其在临床上的推广。最新的技术发展是将UTE技术和指纹成像技术MRF结合,充分利用后者的快速成像和多个激发脉冲性质,将3维全脑髓鞘大分子成像成功压缩至15 min内(Li等,2019, 2020)。
1.3 扩散参数的数据异质性研究
测量间的可比较性是定量成像的重要特征。然而,实际成像时,由于受到成像硬件、序列实现、重建方法和随机噪声等多重因素干扰,定量成像的多次结果间存在不可避免的异质性。当采用不同厂家不同型号的混合研究时,定量结果的异质性更大。
定量成像指标的可重复性评价和校正是当下受到高度重视的关键技术问题(Smith和Nichols,2018)。例如,关注大脑形态学指标,如脑区体积、皮层厚度和面积、皮层曲率等,在多中心测量时存在不可忽略的差异(Glatard等,2015;Schnack等,2010)。磁共振扩散成像反映微观尺度下水分子的扩散行为,可以导出一系列微观结构的模型参数。临床应用较为广泛的是扩散张量成像(diffusion tensor imaging, DTI)模型,White等人(2011)在4个中心使用3台1.5T和1台3T MRI扫描了超过100例精神分裂症患者和正常被试,在两类人群多个脑区的各向异性分数(fractional anisotropy)中均发现了与中心相关的差异。特别地,在其中一台1.5 T扫描仪上,所统计的病人和正常人之间的差异与其他中心差别较大,甚至大于不同场强扫描仪间的差别。即使方差分析中引入不同中心作为协变量,但各中心的影响仍无法消除。这表明简单的线性模型无法准确描述机器间差异。Tong等人(2019)研究了神经密度成像(track density imaging, TDI)在严格一致序列参数下的可重复性,发现不同中心间的数据可重复性仍然低于同一中心内,在不同白质区域间比较,显示单方向纤维分布的区域比多方向混合分布的纤维区域可重复性更高,且多b值采集获得的TDI系数均比单b值的可重复性更高。更高阶模型的扩散峰度成像(diffusional kurtosis imaging, DKI)需要拟合多达21个参数,成像伪影和噪声显著加剧四阶峰度张量的拟合误差,导致模型估计在部分区域不准确甚至出现异常值(Tabesh等,2011)。定量弛豫参数方法的可重复性研究报告相对积极,但其中存在因技术限制相关的局限,还有待进一步更广泛的比较研究。
总体而言,定量磁共振成像受到实际物理因素影响,导致测量可重复性下降的问题不可回避。需要应用校正融合的方法减小数据间的异质性。近年来,多中心数据融合方法研究发展较快。ComBat是目前使用范围较广泛的一种基于统计校正的方案。该方法将不同中心或被试等相关的协变量作为中心效应量并进行校正,可应用在小样本数据的融合中,在显著降低中心间差异的同时保留被试的生理多样性。ComBat及其变体在大脑灰质皮层(Fortin等,2018)、DTI模型(Fortin等,2017)以及脑功能网络(Yu等,2018)等系数的多中心校正中都有应用。近期有研究将迁移学习与ComBat方法结合,用于校正在已知中心的新采集数据,融合包括结构、扩散MRI以及PET等多模态放射组学指标(Da-Ano等,2021)。增加了主成分协变量的ComBat++方法可用于区分大体量数据特征中的因果效应和未知的混合效应,应用在校正阿尔兹海默疾病数据集(the Alzheimer’s disease neuroimaging initiative, ADNI)中(Wachinger等,2021)。而结合了广义相加模型的ComBat-GAM方法可对大体量的脑结构数据进行校正,消除不同数据集之间偏差的同时保留非线性的年龄趋势信息(Pomponio等,2020)。相比于直接在不同中心采集的不同数据集上应用ComBat,使用了旅行者被试数据做校正的TS-ComBat方法能得到测量偏差更小的校正结果(Maikusa等,2021)。在使用Combat方法校正各中心间的数据特征时,还需对应用的数据集和各个协变量做出合理假设,防止出现矫枉过正的情况(Orlhac等,2022)。
深度学习或机器学习方法为多中心数据融合是一种更为有效的途径。例如,图像质量迁移方法使用了基于数据块回归,即从输入的低质量图像的3维邻域数据块中学习到输出高质量图像中相应体素的映射。该方法在图像超分辨、从单b值扩散成像数据中预测其他b值数据中皆有不错的表现(Alexander等,2017)。Tax等人(2019)比较了4种深度学习方法及1种自适应字典学习的方法在扩散成像数据融合中的表现。结果发现不同的网络各有优劣,但都能较好地减少中心间差异。另一种基于3维分层卷积神经网络的融合方法,以一台高质量成像设备的高阶DKI系数特征为参考,可从其他设备采集的扩散图像中重建出与之相似的高质量DKI特征,显著提高了各项特征指标的中心间一致性以及可重复性(Tong等,2020)。
2 定量PET成像
正电子发射断层成像(PET)(Vaquero和Kinahan,2015)是一种核医学分子影像技术,以放射性核素标记的化合物作为示踪剂,实现体内生理过程和代谢功能的可视化。PET图像显示了不同部位对放射性示踪剂的摄取情况。临床上主要采用静态PET成像以及半定量分析。在示踪剂注射一段时间后,进行10~40 min扫描,并重建单帧静态PET图像。半定量分析指标可以是放射性浓度值、标准化摄取值(standardized uptake value, SUV)(Thie,2004)或SUV比值(SUV ratio, SUVR)。SUV由放射性浓度值关于全身平均摄入剂量标准化得到,能够消除由示踪剂剂量、体重导致的个体放射性浓度差异。采用参考组织SUV校正后的SUVR可以减少因成像系统性能、图像重建方法和噪声水平等因素导致的不同扫描间的差异。
基于模型的定量分析能够对主要生理因素进行量化和区分。示踪剂在某处的摄取量由多种局部生理因素决定,例如血流、示踪剂与血液中生物分子的结合、示踪剂与组织中生物分子的非特异性结合(nonspecific binding)、组织中靶分子的浓度、示踪剂与靶分子的特异性结合(specific binding)和血浆清除率(Carson,2005)。不同示踪剂具有独特的分子性质和代谢路径,因而影响其摄取量的具体生理因素和影响程度也不同。代谢动力学模型描述了生理参数与示踪剂摄取量的关系。
2.1 房室模型
房室模型(compartment model)(Bentourkia和Zaidi,2007)是目前应用最广泛的动力学模型,其中的不同房室代表示踪剂在组织中的不同代谢状态。该模型的一般表达式为
| $ \frac{\mathrm{d} \boldsymbol{C}(t)}{\mathrm{d} t}=\boldsymbol{M C}(t)+\boldsymbol{b} \boldsymbol{C}_{\mathrm{P}}(t) $ | (1) |
式中,${\mathit{\boldsymbol{C}}}$($t$)为各个房室的时间—放射性曲线(time-activity curve, TAC),${\mathit{\boldsymbol{C}}}_{\rm {P}}$($t$)为血浆TAC,即模型的输入函数(input function)。模型参数为速率常数(rate constant),表示单位时间内从一个房室流向另一个房室的浓度的比例,其具体生理意义由其连接的两个房室决定。${\mathit{\boldsymbol{M}}}$和${\mathit{\boldsymbol{b}}}$是由速率常数组成的矩阵。模型输出为所有房室的放射性浓度总和。
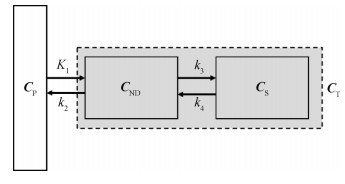
如图 2所示的二房室(two-tissue compartment, 2TC)模型(Gunn等,2015)表示为
| $ \begin{array}{*{20}{c}} {\frac{{{\rm{d}}{\mathit{\boldsymbol{C}}_{{\rm{ND}}}}(t)}}{{{\rm{d}}t}} = {K_1}{\mathit{\boldsymbol{C}}_{\rm{P}}}(t) - }\\ {\left({{k_2} + {k_3}} \right){\mathit{\boldsymbol{C}}_{{\rm{ND}}}}(t) + {k_4}{\mathit{\boldsymbol{C}}_{\rm{S}}}(t)} \end{array} $ | (2) |
| $ \frac{\mathrm{d} \boldsymbol{C}_{\mathrm{S}}(t)}{\mathrm{d} t}=k_{3} \boldsymbol{C}_{\mathrm{ND}}(t)-k_{4} \boldsymbol{C}_{\mathrm{S}}(t) $ | (3) |
| $ \boldsymbol{C}_{\mathrm{T}}(t)=\boldsymbol{C}_{\mathrm{ND}}(t)+\boldsymbol{C}_{\mathrm{S}}(t) $ | (4) |
式中,${\mathit{\boldsymbol{C}}}_{\rm {ND}}$($t$)为不可替换房室的放射性浓度,${\mathit{\boldsymbol{C}}}_{\rm {S}}$($t$)为特异性结合房室的放射性浓度,${\mathit{\boldsymbol{C}}}_{\rm {T}}(t)$为所有房室的放射性浓度,$K_{\rm {1}}$、$k_{\rm {2}}$、$k_{\rm {3}}$、$k_{\rm {4}}$为速率常数。示踪剂在组织中的两种状态通过不可替换房室(non-displaceable compartment)和特异性结合房室(specifically bound compartment)加以区分。
动力学分析通过估计模型参数实现生理参数的量化。动态PET图像是将原始信号划分为多个时间段并重建得到的图像序列,从中可提取感兴趣区域(region of interest, ROI)水平或体素水平的TAC,记为$PET$($t$)。测得$PET$($t$)与${\mathit{\boldsymbol{C}}_{\rm{P}}}(t)$之后,以模型输出${\mathit{\boldsymbol{C}}}_{\rm {T}}$($t$)拟合$PET$($t$)可以得到模型参数的估计。对所有体素的TAC进行拟合还将得到定量的参数图像(Gallezot等,2020)。基于房室模型,不同的参数估计方式对应形成了不同的定量分析方法,例如非线性最小二乘(nonlinear least square, NLS)方法和参考组织法(reference tissue method)(Lammertsma和Hume,1996;Hume等,1992)。
2.2 PET定量成像的误差分析
2.2.1 模型选择
示踪剂在体内的代谢路径十分复杂,涉及多种生理因素。而动力学建模只是对真实情况的近似,无法囊括所有可能的因素。不同的模型复杂度会导致不同程度的偏差。为使参数估计值在统计上更稳定,应选择合理且简单的模型,避免过拟合。因此,建模时往往只考虑1~3种最主要的生理因素。另一方面,若真实生理状态或测量数据不符合模型假设,也将对定量分析结果造成偏差。房室模型一般假设每个房室内部浓度均匀、扫描期间生理参数稳定不变等。参考组织法对于示踪剂在参考组织中的代谢特点也有严格的限制。
合适的定量分析模型应具有以下特点:1)对测量数据拟合度好;2)参数估计值在生理上合理(Ikonomovic等,2008);3)同一个体的生理参数具有较高的重测重现性(test-retest reproducibility) (Cruz等,2020);4)同一人群(例如健康人群、患者)的生理参数具有较低的变异性;5)对主要生理因素的变化敏感,对无关生理因素的扰动以及偏离假设的情形不敏感;6)参数估计方法的计算效率高、PET扫描协议简单。
此外,PET图像显示的放射性浓度可能部分来自组织周围的血管,从中提取的TAC与真实的组织TAC存在一定偏差,尤其是对于血管分布较多或靠近心室的组织。针对这一现象,可以根据组织中血管体积的占比VB,将组织和血管中的放射性浓度加权求和,作为模型输出。VB可以取经验值,例如5%,也可以作为参数和其他模型参数同时进行估计。
2.2.2 图像质量
PET定量分析的准确性极易受图像质量的影响。受探测器晶体宽度、正电子射程(positron range)和伽马光子非共线性(photon non-collinearity)等因素的限制,成像系统的空间分辨率较低(Moses,2011)。图像重建、衰减校正(attenuation correction)、散射校正(scatter correction)和图像平滑等过程的方法选择也将决定最终的图像质量。噪声水平与探测器晶体灵敏度、示踪剂注射剂量和扫描时长等因素有关。特别是在动态PET成像中,早期图像帧持续时间短、信噪比低以及散射校正难度大。由于PET图像分辨率较低,小ROI或体素水平的分析存在部分容积效应(partial volume effect)。为提高定量分析的准确性,需要对该效应进行相应的校正(Erlandsson等,2012)。
衰减校正和ROI选取需要借助计算机断层成像(computed tomography, CT)或磁共振图像中的信息。因此,结构图像与PET图像的配准情况也将间接影响参数估计的准确性。被试在扫描期间的运动容易造成PET图像模糊,且增加了与结构图像配准的难度。动态图像的逐帧对齐(frame-to-frame alignment)是PET脑成像中最常用的头动校正方法,但该方法对帧内运动无法校正。此外,肺、心脏和肠胃等器官的运动更复杂,需要专门的校正方法。
2.2.3 输入函数测量误差
PET定量分析一般要求测量输入函数,具体是在动态PET扫描期间对桡动脉(radial artery)进行多次抽血,并通过拟合模型(Feng等,1993)或插值得到输入函数。动脉抽血具有侵入性且操作复杂,血液样本的后续处理、测量与校正也较为烦琐,容易引入误差,进而影响定量分析的准确性(Chen等,1991)。
为了简化输入函数的获取方式,研究者们提出了几类输入函数估计方法,主要有图像获取输入函数(image-derived input function, IDIF)(Zanotti-Fregonara等,2011)、基于群体的输入函数(population-based input function, PBIF)(Buchert等,2020)以及与动力学参数同时估计的输入函数(simultaneous estimation of the input function, SIME)(Feng等,2020)。IDIF和PBIF方法分别对个体PET图像中获取的血管TAC和群体输入函数模板进行校正,得到个体的输入函数估计。为得到更准确的校正系数,仍然需要抽取少数血液样本。SIME方法首先建立输入函数的解析模型,随后通过拟合多个ROI的TAC对输入函数参数和各个ROI的动力学参数同时进行估计。为了同时确定速率常数K1和输入函数的幅值,SIME方法至少需要一次动脉抽血。IDIF方法基于图像估计输入函数,因此容易受到前文所述的部分容积效应、图像噪声等因素以及图像配准情况的影响。尤其在脑成像中,颈动脉等血管的直径较小,IDIF方法应用效果较差。
2.3 最新进展
PET设备、图像重建方法和图像处理方法的最新进展使PET图像质量大幅提升,间接提高了定量动力学分析和参数成像的准确性。
硬件设备方面,目前新一代飞行时间PET (time-of-flight PET, TOF-PET)(Surti和Karp,2016)已实现约200 ps的符合时间分辨率(coincidence timing resolution),能够将湮灭事件的定位缩小至3 cm以内,极大地改善了图像质量。PET/MRI系统(Vandenberghe和Marsden,2015)可以同时采集PET和MRI两种模态的图像,不需要再进行配准,有利于准确定位ROI、估计输入函数(Zanotti-Fregonara等,2011)和运动校正(Catana等,2011)。全身PET (total-body PET)(Badawi等,2019;Cherry等,2018;Pantel等,2020)具有超长轴向视野和超高灵敏度。全球首台全身PET/CT系统uEXPLORER(Badawi等,2019)的轴向视野长达1.94 m,系统灵敏度实现了40倍提升。研究表明uEXPLORER在低剂量成像(Liu等,2021a; Hu等,2021)、快速成像(Zhang等,2020a)(Liu等,2021b)和长时间延迟成像(Hu等,2021)方面具有显著优势。高质量图像使定量分析的参数估计结果更稳定。同时,基于全身PET/CT系统的动态成像和定量分析通过心脏、主动脉等部位的TAC估计输入函数,可以避免动脉抽血。目前已有研究基于uEXPLORER和[18F] 氟脱氧葡萄糖(fluorodeoxyglucose, FDG)实现了全身葡萄糖代谢定量成像(Zhang等,2020b)。
人工智能和深度学习方法广泛应用于解决各类医学图像问题。对于PET成像,深度神经网络在信号处理、衰减和散射校正(Yang等,2019)、图像重建(Reader等,2021)、去噪(Cui等,2019)、超分(Song等,2020)以及配准方面均有应用(Visvikis等,2019),从多个方面改善了PET图像质量。
关于PET定量分析方法,传统动力学模型假设参数在扫描期间为常数,即假设生理状态稳定不变。最近有研究人员提出了参数随时间变化的模型,用于测量神经递质水平的变化(Ceccarini等,2020)。前文所述的基于动态PET图像进行体素水平定量分析的过程称为间接参数成像。与此相对,直接参数成像将定量模型与图像重建结合,直接从投影数据重建参数图像(Gallezot等,2020)。此外,机器学习和深度学习也已应用于输入函数估计(Kuttner等,2020)和生理参数估计中(Pan等,2017;Wang等,2019)。
3 结语
大脑定量成像技术发展需要面向临床诊断的需求,探索生理意义明确且可靠的定量指标。如何进一步实现高效率、高精度和高稳定地获取成像定量信息,仍然是当前技术研究的重要突破方向。
MRI定量参数成像应实现从单个参数到多个参数同时定量、从表观参数到微观参数、从实验室到临床的转变。这一过程仍然依赖MRI成像系统硬件的提升,仍然依赖配套的成像序列在速度、精准度等方面的优化,仍然依赖成像配套的微观模型的建立、后处理和显示软件的提升。在得到稳定可靠的定量参数的基础上,后续工作还应探索如何将定量参数结果做标准化的诊断工作。
PET技术应结合合适的示踪剂、复杂定量分析方法和MRI等多模态成像,提供更丰富、更精准的代谢信息(Meikle等,2021)。定量模型需要考虑示踪剂在全身范围内的传输、不同部位之间输入函数的差异和不同器官的生理特点。这个过程中PET成像系统和图像处理方法的发展非常重要。比如进一步提升TOF-PET的符合时间分辨率(Lecoq,2017)、研制更高分辨率的脑部专用PET成像系统以及尝试新的晶体材料以降低PET设备的成本等等。对于PET图像处理与分析,后续工作应探索如何基于深度学习方法形成数据采集和处理的标准化流程,以及如何在临床上应用和验证深度学习方法。
最后,随着PET/MRI成像设备的日趋成熟和推广,PET/MR同时定量将是一个重要的发展方向。通过同时成像时匹配的位置信息、运动信息,联合重建以同时定量多项生理、物理参数,从而获得对组织信息更全面的表征。
致谢 本文由中国图象图形学学会医学影像专业委员会组织撰写,该专委会更多详情请见链接:http://www.csig.org.cn/detail/2388。
参考文献
-
Alexander D C, Zikic D, Ghosh A, Tanno R, Wottschel V, Zhang J Y, Kaden E, Dyrby T B, Sotiropoulos S N, Zhang H, Criminisi A. 2017. Image quality transfer and applications in diffusion MRI. Neuroimage, 152: 283-298 [DOI:10.1016/j.neuroimage.2017.02.089]
-
Badawi R D, Shi H C, Hu P C, Chen S G, Xu T Y, Price P M, Ding Y, Spencer B A, Nardo L, Liu W P, Bao J, Jones T, Li H D, Cherry S R. 2019. First human imaging studies with the EXPLORER total-body PET scanner. The Journal of Nuclear Medicine, 60(3): 299-303 [DOI:10.2967/jnumed.119.226498]
-
Baranovicova E, Mlynarik V, Kantorova E, Hnilicova P, Dobrota D. 2016. Quantitative evaluation of cerebral white matter in patients with multiple sclerosis using multicomponent T2 mapping. Neurological Research, 38(5): 389-396 [DOI:10.1080/01616412.2016.1165450]
-
Bentourkia M, Zaidi H. 2007. Tracer kinetic modeling in PET. PET Clinics, 2(2): 267-277 [DOI:10.1016/J.CPET.2007.08.003]
-
Bernstein M A, King K F, Zhou X J. 2004. Handbook of MRF Pulse Sequences. Boston: Academic Press
-
Björk M, Zachariah D, Kullberg J, Stoica P. 2016. A multicomponent T2 relaxometry algorithm for myelin water imaging of the brain. Magnetic Resonance in Medicine, 75(1): 390-402 [DOI:10.1002/mrm.25583]
-
Buchert R, Dirks M, Schütze C, Wilke F, Mamach M, Wirries A K, Pflugrad H, Hamann L, Langer L B N, Wetzel C, Lukacevic M, Polyak A, Kessler M, Petrusch C, Bengel F M, Geworski L, Rupprecht R, Weissenborn K, Ross T L, Berding G. 2020. Reliable quantification of 18F-GE-180 PET neuroinflammation studies using an individually scaled population-based input function or late tissue-to-blood ratio. European Journal of Nuclear Medicine and Molecular Imaging, 47(12): 2887-2900 [DOI:10.1007/s00259-020-04810-1]
-
Cao X Z, Liao C Y, Iyer S S, Wang Z X, Zhou Z H, Dai E P, Liberman G, Dong Z J, Gong T, He H J, Zhong J H, Bilgic B and Setsompop K. 2022. Optimized multi-axis spiral projection MR fingerprinting with subspace reconstruction for rapid whole-brain high-isotropic-resolution quantitative imaging. Magnetic Resonance in Medicine: #29194[DOI: 10.1002/mrm.29194]
-
Cao X Z, Ye H H, Liao C Y, Li Q, He H J, Zhong J H. 2019. Fast 3D brain MR fingerprinting based on multi-axis spiral projection trajectory. Magnetic Resonance in Medicine, 82(1): 289-301 [DOI:10.1002/mrm.27726]
-
Carson R E. 2005. Tracer kinetic modeling in PET//Bailey D L, Townsend D W, Valk P E and Maisey M N, eds. Positron Emission Tomography. London: Springer: 127-159[DOI: 10.1007/1-84628-007-9_6]
-
Catana C, Benner T, Van Der Kouwe A, Byars L, Hamm M, Chonde D B, Michel C J, El Fakhri G, Schmand M, Sorensen A G. 2011. MRI-assisted PET motion correction for neurologic studies in an integrated MR-PET scanner. The Journal of Nuclear Medicine, 52(1): 154-161 [DOI:10.2967/jnumed.110.079343]
-
Ceccarini J, Liu H, Van Laere K, Morris E D, Sander C Y. 2020. Methods for quantifying neurotransmitter dynamics in the living brain with PET imaging. Frontiers in Physiology, 11: #792 [DOI:10.3389/fphys.2020.00792]
-
Chen K, Huang S C, Yu D C. 1991. The effects of measurement errors in the plasma radioactivity curve on parameter estimation in positron emission tomography. Physics in Medicine and Biology, 36(9): 1183-1200 [DOI:10.1088/0031-9155/36/9/003]
-
Cherry S R, Jones T, Karp J S, Qi J Y, Moses W W, Badawi R D. 2018. Total-body PET: maximizing sensitivity to create new opportunities for clinical research and patient care. The Journal of Nuclear Medicine, 59(1): 3-12 [DOI:10.2967/jnumed.116.184028]
-
Choi J Y, Hart T, Whyte J, Rabinowitz A R, Oh S H, Lee J, Kim J J. 2019. Myelin water imaging of moderate to severe diffuse traumatic brain injury. NeuroImage: Clinical, 22: #101785 [DOI:10.1016/j.nicl.2019.101785]
-
Christen T, Pannetier N A, Ni W W, Qiu D, Moseley M E, Schuff N, Zaharchuk G. 2014. MR vascular fingerprinting: a new approach to compute cerebral blood volume, mean vessel radius, and oxygenation maps in the human brain. NeuroImage, 89: 262-270 [DOI:10.1016/j.neuroimage.2013.11.052]
-
Cloos M A, Knoll F, Zhao T J, Block K T, Bruno M, Wiggins G C, Sodickson D K. 2016. Multiparametric imaging with heterogeneous radiofrequency fields. Nature Communications, 7(1): #12445 [DOI:10.1038/ncomms12445]
-
Cohen O, Huang S N, McMahon M T, Rosen M S, Farrar C T. 2018. Rapid and quantitative chemical exchange saturation transfer (CEST) imaging with magnetic resonance fingerprinting (MRF). Magnetic Resonance in Medicine, 80(6): 2449-2463 [DOI:10.1002/mrm.27221]
-
Cruz G, Jaubert O, Qi H K, Bustin A, Milotta G, Schneider T, Koken P, Doneva M, Botnar R M, Prieto C. 2020. 3D free-breathing cardiac magnetic resonance fingerprinting. NMR in Biomedicine, 33(10): #4370 [DOI:10.1002/nbm.4370]
-
Cui J N, Gong K, Guo N, Wu C X, Meng X X, Kim K, Zheng K, Wu Z F, Fu L P, Xu B X, Zhu Z H, Tian J H, Liu H F, Li Q Z. 2019. PET image denoising using unsupervised deep learning. European Journal of Nuclear Medicine and Molecular Imaging, 46(13): 2780-2789 [DOI:10.1007/s00259-019-04468-4]
-
Da-Ano R, Lucia F, Masson I, Abgral R, Alfieri J, Rousseau C, Mervoyer A, Reinhold C, Pradier O, Schick U, Visvikis D, Hatt M. 2021. A transfer learning approach to facilitate ComBat-based harmonization of multicentre radiomic features in new datasets. PLoS One, 16(7): #0253653 [DOI:10.1371/journal.pone.0253653]
-
Deoni S C L, Rutt B K, Peters T M. 2003. Rapid combined T1 and T2 mapping using gradient recalled acquisition in the steady state. Magnetic Resonance in Medicine, 49(3): 515-526 [DOI:10.1002/mrm.10407]
-
Erlandsson K, Buvat I, Pretorius P H, Thomas B A, Hutton B F. 2012. A review of partial volume correction techniques for emission tomography and their applications in neurology, cardiology and oncology. Physics in Medicine and Biology, 57(21): R119-R159 [DOI:10.1088/0031-9155/57/21/R119]
-
Faizy T D, Thaler C, Broocks G, Flottmann F, Leischner H, Kniep H, Nawabi J, Schön G, Stellmann J P, Kemmling A, Reddy R, Heit J J, Fiehler J, Kumar D, Hanning U. 2020. The myelin water fraction serves as a marker for age-related myelin alterations in the cerebral white matter——a multiparametric MRI aging study. Frontiers in Neuroscience, 14: #136 [DOI:10.3389/fnins.2020.00136]
-
Feng D D, Chen K W, Wen L F. 2020. Noninvasive input function acquisition and simultaneous estimations with physiological parameters for PET quantification: a brief review. IEEE Transactions on Radiation and Plasma Medical Sciences, 4(6): 676-683 [DOI:10.1109/trpms.2020.3010844]
-
Feng D G, Huang S C, Wang X M. 1993. Models for computer simulation studies of input functions for tracer kinetic modeling with positron emission tomography. International Journal of Bio-Medical Computing, 32(2): 95-110 [DOI:10.1016/0020-7101(93)90049-C]
-
Fortin J P, Cullen N, Sheline Y I, Taylor W D, Aselcioglu I, Cook P A, Adams P, Cooper C, Fava M, McGrath P J, McInnis M, Phillips M L, Trivedi M H, Weissman M M, Shinohara R T. 2018. Harmonization of cortical thickness measurements across scanners and sites. NeuroImage, 167: 104-120 [DOI:10.1016/j.neuroimage.2017.11.024]
-
Fortin J P, Parker D, Tunç B, Watanabe T, Elliott M A, Ruparel K, Roalf D R, Satterthwaite T D, Gur R C, Gur R E, Schultz R T, Verma R, Shinohara R T. 2017. Harmonization of multi-site diffusion tensor imaging data. NeuroImage, 161: 149-170 [DOI:10.1016/j.neuroimage.2017.08.047]
-
Gallezot J D, Lu Y H, Naganawa M, Carson R E. 2020. Parametric Imaging with PET and SPECT. IEEE Transactions on Radiation and Plasma Medical Sciences, 4(1): 1-23 [DOI:10.1109/TRPMS.2019.2908633]
-
Glatard T, Lewis L B, Da Silva R F, Adalat R, Beck N, Lepage C, Rioux P, Rousseau M E, Sherif T, Deelman E, Khalili-Mahani N, Evans A C. 2015. Reproducibility of neuroimaging analyses across operating systems. Frontiers in Neuroinformatics, 9: #12 [DOI:10.3389/fninf.2015.00012]
-
Gunn R N, Slifstein M, Searle G E, Price J C. 2015. Quantitative imaging of protein targets in the human brain with PET. Physics in Medicine and Biology, 60(22): 363-411 [DOI:10.1088/0031-9155/60/22/R363]
-
Homer J, Beevers M S. 1985. Driven-equilibrium single-pulse observation of T1 relaxation.A reevaluation of a rapid "new" method for determining NMR spin-lattice relaxation times. Journal of Magnetic Resonance, 63(2): 287-297 [DOI:10.1016/0022-2364(85)90318-X]
-
Hu P C, Lin X, Zhuo W H, Tan H, Xie T W, Liu G B, Chen S G, Chen X, Yu H J, Zhang Y Q, Shi H C, Liu H K. 2021. Internal dosimetry in F-18 FDG PET examinations based on long-time-measured organ activities using total-body PET/CT: does it make any difference from a short-time measurement?. EJNMMI Physics, 8(1): #51 [DOI:10.1186/s40658-021-00395-2]
-
Hume S P, Myers R, Bloomfield P M, Opacka-Juffry J, Cremer J E, Ahier R G, Luthra S K, Brooks D J, Lammertsma A A. 1992. Quantitation of Carbon-11-labeled raclopride in rat striatum using positron emission tomography. Synapse, 12(1): 47-54 [DOI:10.1002/syn.890120106]
-
Ikonomovic M D, Klunk W E, Abrahamson E E, Mathis C A, Price J C, Tsopelas N D, Lopresti B J, Ziolko S, Bi W Z, Paljug W R, Debnath M L, Hope C E, Isanski B A, Hamilton R L, Dekosky S T. 2008. Post-mortem correlates of in vivo PiB-PET amyloid imaging in a typical case of Alzheimer's disease. Brain, 131(6): 1630-1645 [DOI:10.1093/brain/awn016]
-
Kuttner S, Wickstrøm K K, Kalda G, Esmaeil Dorraji S, Martin-Armas M, Oteiza A, Jenssen R, Fenton K, Sundset R, Axelsson J. 2020. Machine learning derived input-function in a dynamic 18F-FDG PET study of mice. Biomedical Physics and Engineering Express, 6(1): #015020 [DOI:10.1088/2057-1976/ab6496]
-
Lammertsma A A, Hume S P. 1996. Simplified reference tissue model for PET receptor studies. NeuroImage, 4(3): 153-158 [DOI:10.1006/NIMG.1996.0066]
-
Lecoq P. 2017. Pushing the limits in time-of-flight PET imaging. IEEE Transactions on Radiation and Plasma Medical Sciences, 1(6): 473-485 [DOI:10.1109/TRPMS.2017.2756674]
-
Li Q, Cao X, Ye H, Zhou Z, He H and Zhong J. 2020.3D UTE-MRF for multiple parameteric maps with sub-millimeter isotropic resolution using multi-dimensional golden-angle radial trajectory//Int Soc Magn Reson Med
-
Li Q, Cao X Z, Ye H H, Liao C Y, He H J, Zhong J H. 2019. Ultrashort echo time magnetic resonance fingerprinting (UTE-MRF) for simultaneous quantification of long and ultrashort T2 tissues. Magnetic Resonance in Medicine, 82(4): 1359-1372 [DOI:10.1002/mrm.27812]
-
Liao C Y, Bilgic B, Manhard M K, Zhao B, Cao X Z, Zhong J H, Wald L L, Setsompop K. 2017. 3D MR fingerprinting with accelerated stack-of-spirals and hybrid sliding-window and GRAPPA reconstruction. NeuroImage, 162: 13-22 [DOI:10.1016/j.neuroimage.2017.08.030]
-
Liao C Y, Wang K, Cao X Z, Li Y P, Wu D C, Ye H H, Ding Q P, He H J, Zhong J H. 2018. Detection of lesions in mesial temporal lobe epilepsy by using MR fingerprinting. Radiology, 288(3): 804-812 [DOI:10.1148/radiol.2018172131]
-
Liu G B, Hu P C, Yu H J, Tan H, Zhang Y Q, Yin H Y, Hu Y, Gu J Y, Shi H C. 2021a. Ultra-low-activity total-body dynamic PET imaging allows equal performance to full-activity PET imaging for investigating kinetic metrics of 18F-FDG in healthy volunteers. European Journal of Nuclear Medicine and Molecular Imaging, 48(8): 2373-2383 [DOI:10.1007/s00259-020-05173-3]
-
Liu G B, Yu H J, Shi D, Hu P C, Hu Y, Tan H, Zhang Y Q, Yin H Y and Shi H C. 2021b. Short-time total-body dynamic PET imaging performance in quantifying the kinetic metrics of 18F-FDG in healthy volunteers. European Journal of Nuclear Medicine and Molecular Imaging[DOI: 10.1007/s00259-021-05500-2]
-
Look D C, Locker D R. 1970. Time saving in measurement of NMR and EPR relaxation times. Review of Scientific Instruments, 41(2): 250-251 [DOI:10.1063/1.1684482]
-
Ma D, Gulani V, Seiberlich N, Liu K C, Sunshine J L, Duerk J L, Griswold M A. 2013. Magnetic resonance fingerprinting. Nature, 495(7440): 187-192 [DOI:10.1038/nature11971]
-
Ma S, Wang N, Fan Z Y, Kaisey M, Sicotte N L, Christodoulou A G, Li D B. 2021. Three-dimensional whole-brain simultaneous T1, T2, and T1ρ quantification using MR Multitasking: Method and initial clinical experience in tissue characterization of multiple sclerosis. Magnetic Resonance in Medicine, 85(4): 1938-1952 [DOI:10.1002/mrm.28553]
-
Ma Y J, Jang H, Wei Z, Cai Z Y, Xue Y P, Lee R R, Chang E Y, Bydder G M, Corey-Bloom J, Du J. 2020a. Myelin imaging in human brain using a short repetition time adiabatic inversion recovery prepared ultrashort echo time (STAIR-UTE) MRI sequence in multiple sclerosis. Radiology, 297(2): 392-404 [DOI:10.1148/RADIOL.2020200425]
-
Ma Y J, Searleman A C, Jang H, Wong J, Chang E Y, Corey-Bloom J, Bydder G M, Du J. 2020b. Whole-brain myelin imaging using 3D double-echo sliding inversion recovery ultrashort echo time (DESIRE UTE) MRI. Radiology, 294(2): 362-374
-
Maikusa N, Zhu Y H, Uematsu A, Yamashita A, Saotome K, Okada N, Kasai K, Okanoya K, Yamashita O, Tanaka S C, Koike S. 2021. Comparison of traveling-subject and ComBat harmonization methods for assessing structural brain characteristics. Human Brain Mapping, 42(16): 5278-5287 [DOI:10.1002/hbm.25615]
-
Meikle S R, Sossi V, Roncali E, Cherry S R, Banati R, Mankoff D, Jones T, James M, Sutcliffe J, Ouyang J S, Petibon Y, Ma C, El Fakhri G, Surti S, Karp J S, Badawi R D, Yamaya T, Akamatsu G, Schramm G, Rezaei A, Nuyts J, Fulton R, Kyme A, Lois C, Sari H, Price J, Boellaard R, Jeraj R, Bailey D L, Eslick E, Willowson K P, Dutta J. 2021. Quantitative PET in the 2020 s: a roadmap. Physics in Medicine and Biology, 66(6): #06RM01 [DOI:10.1088/1361-6560/abd4f7]
-
Moses W W. 2011. Fundamental limits of spatial resolution in PET. Nuclear Instruments and Methods in Physics Research Section A: Accelerators, Spectrometers, Detectors and Associated Equipment, 648(S1): 236-240 [DOI:10.1016/J.NIMA.2010.11.092]
-
Nagtegaal M, Koken P, Amthor T, De Bresser J, Mädler B, Vos F, Doneva M. 2020. Myelin water imaging from multi-echo T2 MR relaxometry data using a joint sparsity constraint. NeuroImage, 219: #117014 [DOI:10.1016/j.neuroimage.2020.117014]
-
Orlhac F, Eertink J J, Cottereau A S, Zijlstra J M, Thieblemont C, Meignan M A, Boellaard R, Buvat I. 2022. A guide to ComBat harmonization of imaging biomarkers in multicenter studies. The Journal of Nuclear Medicine, 63(2): 172-179 [DOI:10.2967/jnumed.121.262464]
-
Pan L Y, Cheng C X, Haberkorn U, Dimitrakopoulou-Strauss A. 2017. Machine learning-based kinetic modeling: a robust and reproducible solution for quantitative analysis of dynamic PET data. Physics in Medicine and Biology, 62(9): 3566-3581 [DOI:10.1088/1361-6560/aa6244]
-
Pantel A R, Viswanath V, Daube-Witherspoon M E, Dubroff J G, Muehllehner G, Parma M J, Pryma D A, Schubert E K, Mankoff D A, Karp J S. 2020. PennPET explorer: human imaging on a whole-body imager. The Journal of Nuclear Medicine, 61(1): 144-151 [DOI:10.2967/jnumed.119.231845]
-
Pomponio R, Erus G, Habes M, Doshi J, Srinivasan D, Mamourian E, Bashyam V, Nasrallah I M, Satterthwaite T D, Fan Y, Launer L J, Masters C L, Maruff P, Zhuo C J, Völzke H, Johnson S C, Fripp J, Koutsouleris N, Wolf D H, Gur R, Gur R, Morris J, Albert M S, Grabe H J, Resnick S M, Nick Bryan R, Wolk D A, Shinohara R T, Shou H C, Davatzikos C. 2020. Harmonization of large MRI datasets for the analysis of brain imaging patterns throughout the lifespan. NeuroImage, 208: #116450 [DOI:10.1016/j.neuroimage.2019.116450]
-
Reader A J, Corda G, Mehranian A, da Costa-Luis C, Ellis S, Schnabel J A. 2021. Deep learning for PET image reconstruction. IEEE Transactions on Radiation and Plasma Medical Sciences, 5(1): 1-25 [DOI:10.1109/trpms.2020.3014786]
-
Rieger B, Zimmer F, Zapp J, Weingärtner S, Schad L R. 2017. Magnetic resonance fingerprinting using echo-planar imaging: Joint quantification of T1 and T2* relaxation times. Magnetic Resonance in Medicine, 78(5): 1724-1733 [DOI:10.1002/mrm.26561]
-
Schmitt P, Griswold M A, Jakob P M, Kotas M, Gulani V, Flentje M, Haase A. 2004. Inversion recovery TrueFISP: quantification of T1, T2, and spin density. Magnetic Resonance in Medicine, 51(4): 661-667 [DOI:10.1002/mrm.20058]
-
Schnack H G, Van Haren N E M, Brouwer R M, Van Baal G C M, Picchioni M, Weisbrod M, Sauer H, Cannon T D, Huttunen M, Lepage C, Collins D L, Evans A, Murray R M, Kahn R S, Hulshoff Pol H E. 2010. Mapping reliability in multicenter MRI: Voxel-based morphometry and cortical thickness. Human Brain Mapping, 31(12): 1967-1982 [DOI:10.1002/hbm.20991]
-
Shao H, Chang E Y, Pauli C, Zanganeh S, Bae W, Chung C B, Tang G, Du J. 2016. UTE bi-component analysis of T2* relaxation in articular cartilage. Osteoarthritis and Cartilage, 24(2): 364-373 [DOI:10.1016/j.joca.2015.08.017]
-
Sheth V, Shao H D, Chen J, Vandenberg S, Corey-Bloom J, Bydder G M, Du J. 2016. Magnetic resonance imaging of myelin using ultrashort echo time (UTE) pulse sequences: phantom, specimen, volunteer and multiple sclerosis patient studies. NeuroImage, 136: 37-44 [DOI:10.1016/j.neuroimage.2016.05.012]
-
Smith S M, Nichols T E. 2018. Statistical challenges in "Big Data" human neuroimaging. Neuron, 97(2): 263-268 [DOI:10.1016/j.neuron.2017.12.018]
-
Song T A, Chowdhury S R, Yang F, Dutta J. 2020. PET image super-resolution using generative adversarial networks. Neural Networks, 125: 83-91 [DOI:10.1016/j.neunet.2020.01.029]
-
Su P, Mao D, Liu P Y, Li Y, Pinho M C, Welch B G, Lu H Z. 2017. Multiparametric estimation of brain hemodynamics with MR fingerprinting ASL. Magnetic Resonance in Medicine, 78(5): 1812-1823 [DOI:10.1002/mrm.26587]
-
Surti S, Karp J S. 2016. Advances in time-of-flight PET. Physica Medica, 32(1): 12-22 [DOI:10.1016/J.EJMP.2015.12.007]
-
Tabesh A, Jensen J H, Ardekani B A, Helpern J A. 2011. Estimation of tensors and tensor-derived measures in diffusional kurtosis imaging. Magnetic Resonance in Medicine, 65(3): 823-836 [DOI:10.1002/mrm.22655]
-
Tax C M, Grussu F, Kaden E, Ning L P, Rudrapatna U, John Evans C, St-Jean S, Leemans A, Koppers S, Merhof D, Ghosh A, Tanno R, Alexander D C, Zappalà S, Charron C, Kusmia S, Ej Linden D, Jones D K, Veraart J. 2019. Cross-scanner and cross-protocol diffusion MRI data harmonisation: a benchmark database and evaluation of algorithms. NeuroImage, 195: 285-299 [DOI:10.1016/j.neuroimage.2019.01.077]
-
Thie J A. 2004. Understanding the standardized uptake value, its methods, and implications for usage. The Journal of Nuclear Medicine, 45(9): 1431-1434
-
Tong Q Q, Gong T, He H J, Wang Z, Yu W W, Zhang J J, Zhai L H, Cui H S, Meng X, Tax C W M, Zhong J H. 2020. A deep learning-based method for improving reliability of multicenter diffusion kurtosis imaging with varied acquisition protocols. Magnetic Resonance Imaging, 73: 31-44 [DOI:10.1016/j.mri.2020.08.001]
-
Tong Q Q, He H J, Gong T, Li C, Liang P P, Qian T Y, Sun Y, Ding Q P, Li K C, Zhong J H. 2019. Reproducibility of multi-shell diffusion tractography on traveling subjects: a multicenter study prospective. Magnetic Resonance Imaging, 59: 1-9 [DOI:10.1016/j.mri.2019.02.011]
-
Vandenberghe S, Marsden P K. 2015. PET-MRI: a review of challenges and solutions in the development of integrated multimodality imaging. Physics in Medicine and Biology, 60(4): 115-154 [DOI:10.1088/0031-9155/60/4/R115]
-
Vaquero J J, Kinahan P. 2015. Positron emission tomography: current challenges and opportunities for technological advances in clinical and preclinical imaging systems. Annual Review of Biomedical Engineering, 17(1): 385-414 [DOI:10.1146/annurev-bioeng-071114-040723]
-
Visvikis D, Cheze Le Rest C, Jaouen V, Hatt M. 2019. Artificial intelligence, machine (Deep) learning and Radio(Geno)Mics: definitions and nuclear medicine imaging applications. European Journal of Nuclear Medicine and Molecular Imaging, 46(13): 2630-2637 [DOI:10.1007/s00259-019-04373-w]
-
Wachinger C, Rieckmann A, Pölsterl S. 2021. Detect and correct bias in multi-site neuroimaging datasets. Medical Image Analysis, 67: #101879 [DOI:10.1016/j.media.2020.101879]
-
Wang B, Ruan D S and Liu H F. 2019. Noninvasive estimation of macro-parameters for dynamic PET images using deep learning//2019 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC). Manchester, UK: IEEE: 1-4[DOI: 10.1109/NSS/MIC42101.2019.9059691]
-
Warntjes J B M, Dahlqvist Leinhard O, West J, Lundberg P. 2008. Rapid magnetic resonance quantification on the brain: Optimization for clinical usage. Magnetic Resonance in Medicine, 60(2): 320-329 [DOI:10.1002/mrm.21635]
-
White T, Magnotta V A, Bockholt H J, Williams S, Wallace S, Ehrlich S, Mueller B A, Ho B C, Jung R E, Clark V P, Lauriello J, Bustillo J R, Schulz S C, Gollub R L, Andreasen N C, Calhoun V D, Lim K O. 2011. Global white matter abnormalities in schizophrenia: a multisite diffusion tensor imaging study. Schizophrenia Bulletin, 37(1): 222-232 [DOI:10.1093/schbul/sbp088]
-
Wilhelm M J, Ong H H, Wehrli S L, Li C, Tsai P H, Hackney D B, Wehrli F W. 2012. Direct magnetic resonance detection of myelin and prospects for quantitative imaging of myelin density. Proceedings of the National Academy of Sciences of the United States of America, 109(24): 9605-9610 [DOI:10.1073/pnas.1115107109]
-
Yang J, Park D, Gullberg G T, Seo Y. 2019. Joint correction of attenuation and scatter in image space using deep convolutional neural networks for dedicated brain 18F-FDG PET. Physics in Medicine and Biology, 64(7): #075019 [DOI:10.1088/1361-6560/ab0606]
-
Ye H H, Cauley S F, Gagoski B, Bilgic B, Ma D, Jiang Y, Du Y P, Griswold M A, Wald L L, Setsompop K. 2017. Simultaneous multislice magnetic resonance fingerprinting (SMS-MRF) with direct-spiral slice-GRAPPA (ds-SG) reconstruction. Magnetic Resonance in Medicine, 77(5): 1966-1974 [DOI:10.1002/mrm.26271]
-
Ye H H, Ma D, Jiang Y, Cauley S F, Du Y P, Wald L L, Griswold M A, Setsompop K. 2016. Accelerating magnetic resonance fingerprinting (MRF) using t-blipped simultaneous multislice (SMS) acquisition. Magnetic Resonance in Medicine, 75(5): 2078-2085 [DOI:10.1002/mrm.25799]
-
Yu M C, Linn K A, Cook P A, Phillips M L, McInnis M, Fava M, Trivedi M H, Weissman M M, Shinohara R T, Sheline Y I. 2018. Statistical harmonization corrects site effects in functional connectivity measurements from multi-site fMRI data. Human Brain Mapping, 39(11): 4213-4227 [DOI:10.1002/hbm.24241]
-
Zanotti-Fregonara P, Chen K W, Liow J S, Fujita M, Innis R B. 2011. Image-derived input function for brain PET studies: Many challenges and few opportunities. Journal of Cerebral Blood Flow and Metabolism, 31(10): 1986-1998 [DOI:10.1038/jcbfm.2011.107]
-
Zhang X Z, Cherry S R, Xie Z H, Shi H C, Badawi R D, Qi J Y. 2020a. Subsecond total-body imaging using ultrasensitive positron emission tomography. Proceedings of the National Academy of Sciences of the United States of America, 117(5): 2265-2267 [DOI:10.1073/pnas.1917379117]
-
Zhang X Z, Xie Z H, Berg E, Judenhofer M S, Liu W P, Xu T Y, Ding Y, Lv Y, Dong Y, Deng Z L, Tang S S, Shi H C, Hu P C, Chen S G, Bao J, Li H D, Zhou J, Wang G B, Cherry S R, Badawi R D, Qi J Y. 2020b. Total-body dynamic reconstruction and parametric imaging on the UEXPLORER. The Journal of Nuclear Medicine, 61(2): 285-291 [DOI:10.2967/jnumed.119.230565]
-
Zhou Z W, Han P, Zhou B, Christodoulou A G, Shaw J L, Deng Z X, Li D B. 2018. Chemical exchange saturation transfer fingerprinting for exchange rate quantification. Magnetic Resonance in Medicine, 80(4): 1352-1363 [DOI:10.1002/mrm.27363]